DataBase
This page provides a list of data sets generated from our research projects that can be viewed and downloaded. Some of the data has been published, while some not yet. If you are interested in using them for further global analysis please firstly contact us Dr. Yaou Shen . We will be pleased to share the data for any specific gene or region analysis.
Repositories
Genome Assembly
-
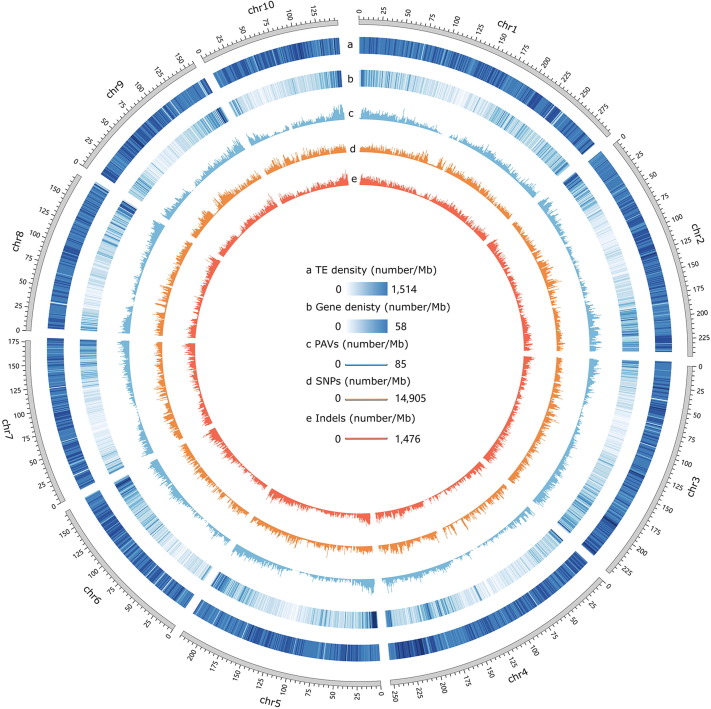
A188 Assemblyed Sequence The current assembled maize genomes cannot represent the broad genetic diversity of maize germplasms. Acquiring more genome sequences is critical for constructing a pan-genome and elucidating the linkage between genotype and phenotype in maize. Here we describe the genome sequence and annotation of A188, a maize inbred line with high phenotypic variation relative to other lines, acquired by single-molecule sequencing and optical genome mapping. We assembled a 2210-Mb genome with a scaffold N50 size of 11.61 million bases (Mb), compared to 9.73 Mb for B73 and 10.2 Mb for Mo17. Based on the B73_RefGen_V4 genome, 295 scaffolds (2084.35 Mb, 94.30% of the final genome assembly) were anchored and oriented on ten chromosomes. Comparative analysis revealed that ~30% of the predicted A188 genes showed large structural divergence from B73, Mo17, and W22 genomes, which causes high protein divergence and may lead to phenotypic variation among the four inbred lines. As a line with high embryonic callus (EC) induction capacity, A188 provides a convenient tool for elucidating the molecular mechanism underlying the formation of EC in maize. Combining our new A188 genome with previously reported QTL and RNA sequencing data revealed eight genes with large structural variation and two differentially expressed genes playing potential roles in maize EC induction.
-
A188 Gene Annotations A total of 80.70% of the A188 genome sequence was identified as transposable-element sequences, including retrotransposons (71.93%), DNA transposons (5.91%), and unclassified elements (2.49%) (Table S5), which was lower than those in B73, Mo17, W22, SK and K0326Y genomes. For retrotransposons, the families of Copia and Gypsy represented respectively 24.01% and 46.92% of the A188 genome (Table S5). For DNA transposons, the representation of the hAT family was much lower than those in the B73 and Mo17 genomes.
Notes: Please contact us for the extraction code!