Germplasm
A total of 362 diverse inbred lines from the current Southwest China breeding program were assembled into a panel. The panel was comprised of 33 parents of major expanded hybrids from Southwest China, 252 new selected and improved inbred lines, 40 representative inbred lines of temperate heterotic groups (Reid, SPT, Lancaster, LRC, etc.) and 37 CIMMYT or U.S. exotic inbred lines. Approximately 17 pairs of inbred lines were parents of local varieties and most of domestic inbred lines were one parent of authorized hybrids in China.
| Name | Pedigree | K = 2 | K = 3 | K = 4 | K = 6 | ||||||||||||||
| Tropical | Temperate | Tropical | NSS | SS | Tropical | PA | PB | Reid | Tropical | PA | PB | Reid | BSSS | North | |||||
| F19 | Derived from CIMMYT germplasm | 0.8442 | 0.1558 | 0.8044 | 0.0302 | 0.1654 | 0.7686 | 0.1012 | 0.0268 | 0.1034 | 0.7301 | 0.0534 | 0.0164 | 0.001 | 0.1979 | 0.0012 | |||
| F06 | Derived from suwan germplasm | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | |||
| Zheng58 | Ye478 mutant | 0.192 | 0.808 | 0.1078 | 0.0644 | 0.8278 | 0.1474 | 0.0044 | 0.0396 | 0.8086 | 0 | 0 | 0 | 0.5509 | 0 | 0.4491 | |||
| Chang7-2 | Wei95×Huangzaosi//S901 | 0.7312 | 0.2688 | 0.6679 | 0.0028 | 0.3293 | 0.6333 | 0.1498 | 0.0006 | 0.2162 | 0.3363 | 0.0518 | 0.0002 | 0.0004 | 0.0766 | 0.5347 | |||
| S273 | Derived from suwan germplasm | 0.864 | 0.136 | 0.8366 | 0.0524 | 0.111 | 0.7902 | 0.111 | 0.0528 | 0.046 | 0.7063 | 0.0898 | 0.0432 | 0.0022 | 0.0108 | 0.1476 | |||
| Su1611 | Derived from suwan germplasm | 0.862 | 0.138 | 0.8402 | 0.1178 | 0.042 | 0.8328 | 0.0292 | 0.1114 | 0.0266 | 0.7836 | 0.0116 | 0.1054 | 0.003 | 0.0148 | 0.0816 | |||
| YA8201 | Brazilian hybrid AGROLERES1051 | 0.8902 | 0.1098 | 0.8602 | 0.0264 | 0.1134 | 0.8581 | 0.0224 | 0.0128 | 0.1066 | 0.7469 | 0.0006 | 0.0088 | 0.0158 | 0.0484 | 0.1796 | |||
| 5311 | Derived from suwan germplasm | 0.998 | 0.002 | 0.992 | 0.0076 | 0.0004 | 0.9964 | 0.0002 | 0.0018 | 0.0016 | 0.9878 | 0 | 0.0022 | 0.0068 | 0.0016 | 0.0016 | |||
| CTL26 | Derived from suwan germplasm | 0.967 | 0.033 | 0.9418 | 0.0582 | 0 | 0.945 | 0.0002 | 0.0544 | 0.0004 | 0.956 | 0 | 0.0412 | 0.0008 | 0.002 | 0 | |||
| 5Gong | 515 / Gongza-1 | 0.9972 | 0.0028 | 0.9814 | 0.018 | 0.0006 | 0.9834 | 0.0028 | 0.012 | 0.0018 | 0.9568 | 0.0006 | 0.004 | 0.0014 | 0.0002 | 0.037 | |||
| 21A | Derived from Tropical germplasm | 0.9332 | 0.0668 | 0.911 | 0.0518 | 0.0372 | 0.8992 | 0.0236 | 0.043 | 0.0342 | 0.8473 | 0.0078 | 0.039 | 0.0074 | 0.0166 | 0.0818 | |||
| SN8-1-1 | Derived from suwan germplasm | 0.9778 | 0.0222 | 0.9638 | 0.0344 | 0.0018 | 0.9634 | 0.0004 | 0.0194 | 0.0168 | 0.9384 | 0 | 0.015 | 0.0088 | 0.0012 | 0.0366 | |||
| Jiao51 | Guizhou landrace Jiaomaerhuangzao | 0.8222 | 0.1778 | 0.7826 | 0.012 | 0.2054 | 0.7706 | 0.0696 | 0.0044 | 0.1554 | 0.5935 | 0.0142 | 0.0018 | 0.018 | 0.0398 | 0.3327 | |||
| V37 | V37 | 0.5782 | 0.4218 | 0.5108 | 0.0468 | 0.4424 | 0.3474 | 0.4234 | 0.0498 | 0.1794 | 0.1258 | 0.3461 | 0.0238 | 0.0394 | 0.003 | 0.4619 | |||
| Mian7317 | Mian7317 | 0.282 | 0.718 | 0.1902 | 0.1498 | 0.6601 | 0.212 | 0.0406 | 0.1302 | 0.6171 | 0.107 | 0.0078 | 0.1194 | 0.4503 | 0.0682 | 0.2474 | |||
| Mian723 | P953 × 698-3 | 0.2762 | 0.7238 | 0.1416 | 0.0576 | 0.8008 | 0 | 0.8656 | 0.004 | 0.1304 | 0 | 0.8579 | 0.0002 | 0.1419 | 0 | 0 | |||
| 7041-5 | Derived from Yogoslavic hybrid SC704 | 0.57 | 0.43 | 0.4764 | 0.0008 | 0.5228 | 0.4591 | 0.1368 | 0 | 0.4041 | 0.1042 | 0.017 | 0.0002 | 0.004 | 0.2592 | 0.6154 | |||
| NZ013-1 | Neizi013-1-1 | 0.3902 | 0.6098 | 0.2987 | 0.0852 | 0.6161 | 0.1196 | 0.5551 | 0.1038 | 0.2216 | 0 | 0.4467 | 0.0472 | 0.001 | 0.0911 | 0.414 | |||
| Yu561 | Nongda202 × 095 | 0.512 | 0.488 | 0.4401 | 0.1413 | 0.4185 | 0.3479 | 0.2821 | 0.1437 | 0.2263 | 0.0928 | 0.2014 | 0.1312 | 0.0174 | 0.0896 | 0.4676 | |||
| H08-155 | H08-155 | 0.5732 | 0.4268 | 0.487 | 0.0016 | 0.5114 | 0.3516 | 0.3724 | 0.0006 | 0.2754 | 0.0862 | 0.2638 | 0.0002 | 0.0784 | 0.0502 | 0.5212 | |||
| K169R | Derived from American hybrid 3163 | 0.4482 | 0.5518 | 0.3418 | 0.006 | 0.6522 | 0.0884 | 0.7416 | 0.0204 | 0.1496 | 0.089 | 0.6627 | 0.0182 | 0.0142 | 0.2126 | 0.0032 | |||
| Dong156 | Dong156 | 0.6786 | 0.3214 | 0.5988 | 0.0008 | 0.4004 | 0.5535 | 0.1676 | 0 | 0.2789 | 0.2166 | 0.0524 | 0.0002 | 0.0008 | 0.0894 | 0.6407 | |||
| M14 | M14 | 0.969 | 0.031 | 0.942 | 0.058 | 0 | 0.9458 | 0.0002 | 0.0536 | 0.0004 | 0.9566 | 0 | 0.041 | 0.0004 | 0.002 | 0 | |||
| Y8G | Derived from American hybrid Y8G61 | 0.4522 | 0.5478 | 0.3996 | 0.2851 | 0.3153 | 0.2078 | 0.4592 | 0.3066 | 0.0264 | 0.2255 | 0.4318 | 0.2991 | 0.007 | 0.0364 | 0.0002 | |||
| B151 | Derived from Pioneer unknown hybrid | 0.362 | 0.638 | 0.3175 | 0.5582 | 0.1242 | 0.2842 | 0.0908 | 0.5566 | 0.0684 | 0.1895 | 0.0376 | 0.5467 | 0.0004 | 0.1147 | 0.1111 | |||
| 618 | Derived from Pioneer unknown hybrid | 0.5718 | 0.4282 | 0.4908 | 0.0052 | 0.504 | 0.0002 | 0.9998 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |||
| 141 | Derived from Pioneer hybrid 78599 | 0.0408 | 0.9592 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |||
| ShuangM9 | M9 × M9B | 0.957 | 0.043 | 0.9396 | 0.0444 | 0.016 | 0.9372 | 0.0036 | 0.034 | 0.0252 | 0.8824 | 0.0006 | 0.028 | 0.0014 | 0.0254 | 0.0622 | |||
| P953 | Derived from hybrid 95-3 | 0.3132 | 0.6868 | 0.2116 | 0.084 | 0.7045 | 0.079 | 0.4577 | 0.0828 | 0.3805 | 0.0006 | 0.4224 | 0.0354 | 0.3382 | 0 | 0.2034 | |||
| LN8 | 478 improved line | 0.3982 | 0.6018 | 0.3045 | 0.0548 | 0.6407 | 0.2964 | 0.114 | 0.0388 | 0.5508 | 0.1586 | 0.0738 | 0.0264 | 0.3546 | 0.1278 | 0.2588 | |||
| HZ127-7 | Derived from American hybrid Y8G61 | 0.4382 | 0.5618 | 0.3875 | 0.3167 | 0.2957 | 0.2158 | 0.4114 | 0.3344 | 0.0384 | 0.2374 | 0.3867 | 0.3217 | 0.0536 | 0.0004 | 0.0002 | |||
| En1824 | En1824 | 0.123 | 0.877 | 0.0366 | 0.1666 | 0.7968 | 0.0792 | 0.0258 | 0.1338 | 0.7612 | 0.0556 | 0.0238 | 0.1222 | 0.6793 | 0.018 | 0.1012 | |||
| ZYDH381-1 | ZYDH381-1 | 0.279 | 0.721 | 0.2044 | 0.2683 | 0.5273 | 0.112 | 0.2942 | 0.2798 | 0.314 | 0.1186 | 0.2517 | 0.2765 | 0.203 | 0.1462 | 0.004 | |||
| Su95-1 | Ye478 × Ji183 | 0.215 | 0.785 | 0.1224 | 0.0634 | 0.8142 | 0.1606 | 0.019 | 0.0432 | 0.7772 | 0.0588 | 0.008 | 0.0256 | 0.6199 | 0.0306 | 0.2571 | |||
| De2010-1 | De2010-1 | 0.133 | 0.867 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |||
| End28 | 78599 improved lines | 0.345 | 0.655 | 0.2955 | 0.5217 | 0.1828 | 0.2961 | 0.028 | 0.5164 | 0.1595 | 0.1138 | 0.0012 | 0.5015 | 0.0314 | 0.0002 | 0.3519 | |||
| 98009 | Shuikouhuang × Wuerhuang // Zi330 | 0.4942 | 0.5058 | 0.4275 | 0.1048 | 0.4677 | 0.4048 | 0.1397 | 0.0999 | 0.3556 | 0.0016 | 0.0426 | 0.0691 | 0.0374 | 0.0294 | 0.8199 | |||
| Mian715 | Hybrid CGT-15 | 0.553 | 0.447 | 0.4397 | 0.5603 | 0 | 0.4409 | 0.0002 | 0.5589 | 0 | 0.4465 | 0 | 0.5533 | 0 | 0.0002 | 0 | |||
| BS08-565 | BS08-565 | 0.4142 | 0.5858 | 0.319 | 0.009 | 0.672 | 0.1544 | 0.4993 | 0.0072 | 0.3391 | 0.122 | 0.469 | 0.0008 | 0.3301 | 0 | 0.078 | |||
| MDH3-200 | MDH3-200 | 0.282 | 0.718 | 0.1736 | 0.036 | 0.7904 | 0.2278 | 0.0176 | 0.004 | 0.7507 | 0.1104 | 0 | 0.0016 | 0.4862 | 0.2486 | 0.1532 | |||
| 21209 | 21209 | 0.261 | 0.739 | 0.1602 | 0.0406 | 0.7992 | 0.1444 | 0.161 | 0.0286 | 0.666 | 0 | 0.0862 | 0.0116 | 0.3027 | 0.2835 | 0.3161 | |||
| De12 | De12 | 0.2886 | 0.7114 | 0.2366 | 0.5672 | 0.1962 | 0.2133 | 0.0897 | 0.5695 | 0.1275 | 0.0006 | 0.007 | 0.5498 | 0.0004 | 0.0002 | 0.442 | |||
| TD1 | 478 improved line | 0.499 | 0.501 | 0.4166 | 0.0656 | 0.5178 | 0.3612 | 0.2062 | 0.0628 | 0.3698 | 0.2357 | 0.1567 | 0.0548 | 0.1959 | 0.1102 | 0.2467 | |||
| Y75 | Shanzong5 × Zhongzong14C4 | 0.6012 | 0.3988 | 0.548 | 0.2196 | 0.2324 | 0.5155 | 0.1172 | 0.2184 | 0.1488 | 0.3037 | 0.0262 | 0.1994 | 0.0004 | 0.1084 | 0.3619 | |||
| Jing07-4 | Qujing landrace | 0.5336 | 0.4664 | 0.4787 | 0.1818 | 0.3395 | 0.4854 | 0.0492 | 0.1734 | 0.292 | 0.1299 | 0 | 0.1597 | 0.0022 | 0.0664 | 0.6418 | |||
| M89 | K22 improved line | 0.2782 | 0.7218 | 0.1763 | 0.0772 | 0.7465 | 0.1044 | 0.2898 | 0.08 | 0.5258 | 0.0322 | 0.238 | 0.0582 | 0.4107 | 0.088 | 0.1728 | |||
| H127RE | American hybrid Y8G61 | 0.4098 | 0.5902 | 0.3564 | 0.3218 | 0.3218 | 0.1706 | 0.4492 | 0.3412 | 0.039 | 0.1947 | 0.4182 | 0.3333 | 0.0292 | 0.0236 | 0.001 | |||
| MH9 | Derived from Mohuang9 population | 0.7032 | 0.2968 | 0.6597 | 0.1022 | 0.238 | 0.6089 | 0.153 | 0.0964 | 0.1418 | 0.5446 | 0.1265 | 0.0916 | 0.061 | 0.0678 | 0.1086 | |||
| NZ925 | NZ925 | 0.354 | 0.646 | 0.2598 | 0.0082 | 0.732 | 0.2683 | 0.0766 | 0.0006 | 0.6545 | 0.0938 | 0.027 | 0.0002 | 0.475 | 0.0408 | 0.3632 | |||
| BS1074 | BS1074 | 0.4752 | 0.5248 | 0.426 | 0.2053 | 0.3687 | 0.4157 | 0.098 | 0.2028 | 0.2835 | 0.086 | 0.0006 | 0.1964 | 0.004 | 0.0924 | 0.6205 | |||
| Lin-1 | 478 improved line | 0.501 | 0.499 | 0.4166 | 0.0576 | 0.5258 | 0.3564 | 0.2189 | 0.0556 | 0.3692 | 0.2237 | 0.1657 | 0.0476 | 0.1877 | 0.1184 | 0.2569 | |||
| Yu9537 | 095 × S37 | 0.5154 | 0.4846 | 0.4433 | 0.1382 | 0.4185 | 0.3516 | 0.2744 | 0.1398 | 0.2342 | 0.1158 | 0.2022 | 0.1268 | 0.0434 | 0.081 | 0.4309 | |||
| JF52-3 | JF52-3 | 0.365 | 0.635 | 0.2954 | 0.308 | 0.3966 | 0.2457 | 0.1725 | 0.3019 | 0.2799 | 0.0134 | 0.0984 | 0.2921 | 0.0874 | 0.1026 | 0.406 | |||
| S7913 | S7913 | 0.6462 | 0.3538 | 0.5874 | 0.0042 | 0.4084 | 0.5657 | 0.1194 | 0.0008 | 0.3141 | 0.3177 | 0.0378 | 0.0002 | 0.1026 | 0.0714 | 0.4702 | |||
| DH29 | Derived from Pioneer hybrid 78599 | 0.075 | 0.925 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |||
| Mo17 | C103 × 187-2 | 0.42 | 0.58 | 0.3613 | 0.2007 | 0.438 | 0.3315 | 0.1426 | 0.1932 | 0.3327 | 0 | 0.0016 | 0.1053 | 0.0378 | 0 | 0.8553 | |||
| JH96B | Derived from Zhongzong2 | 0.4466 | 0.5534 | 0.3903 | 0.2687 | 0.3409 | 0.3651 | 0.1262 | 0.2669 | 0.2418 | 0.0492 | 0.0128 | 0.2374 | 0.0062 | 0.0404 | 0.6539 | |||
| S5003 | Derived from suwan germplasm | 0.977 | 0.023 | 0.9594 | 0.039 | 0.0016 | 0.955 | 0.0104 | 0.0314 | 0.0032 | 0.9108 | 0.002 | 0.0242 | 0.0014 | 0.0024 | 0.0592 | |||
| 2142 | Foreign hybrid Y7864 | 0.5926 | 0.4074 | 0.5237 | 0.0774 | 0.3989 | 0.2687 | 0.6041 | 0.1106 | 0.0166 | 0.1954 | 0.5293 | 0.0844 | 0.0004 | 0.0578 | 0.1326 | |||
| 205-11 | Foreign hybrid Y7865F2 × Qi205 | 0.4922 | 0.5078 | 0.4166 | 0.0922 | 0.4912 | 0.3789 | 0.182 | 0.0892 | 0.3499 | 0.0314 | 0.0714 | 0.078 | 0.0022 | 0.1702 | 0.6468 | |||
| ZJ-3 | WS527 × 7031 | 0.464 | 0.536 | 0.4068 | 0.352 | 0.2412 | 0.4026 | 0.044 | 0.3428 | 0.2106 | 0.2259 | 0.0012 | 0.3243 | 0.1108 | 0.0002 | 0.3377 | |||
| N54-3 | N54-3 | 0.285 | 0.715 | 0.2128 | 0.278 | 0.5092 | 0.2425 | 0.001 | 0.2534 | 0.5031 | 0.1192 | 0 | 0.2174 | 0.4144 | 0.0002 | 0.2488 | |||
| LSC117 | Derived from Mobai1 | 0.9138 | 0.0862 | 0.8886 | 0.011 | 0.1004 | 0.889 | 0.0152 | 0.002 | 0.0938 | 0.819 | 0.002 | 0.0018 | 0.0536 | 0.005 | 0.1186 | |||
| LSC107 | 87-1 × 86-1 | 0.135 | 0.865 | 0.0726 | 0.5782 | 0.3492 | 0.0454 | 0.1124 | 0.5729 | 0.2693 | 0.0062 | 0.0894 | 0.5636 | 0.1951 | 0.0466 | 0.0992 | |||
| Nan09530 | 095 × Zi330 | 0.7476 | 0.2524 | 0.6775 | 0.0014 | 0.3211 | 0.6309 | 0.1796 | 0.0006 | 0.1888 | 0.2683 | 0.062 | 0.0002 | 0 | 0.0186 | 0.6509 | |||
| 98-3 | 698-3 × 81565 // 698-3 | 0.5348 | 0.4652 | 0.4134 | 0.0008 | 0.5858 | 0.1339 | 0.8409 | 0.0006 | 0.0246 | 0.1068 | 0.7817 | 0.0002 | 0 | 0.1104 | 0.0008 | |||
| M232 | 65232/P6C3 // 65232 | 0.388 | 0.612 | 0.3141 | 0.1478 | 0.5381 | 0.3143 | 0.0962 | 0.1354 | 0.4541 | 0.1472 | 0.0478 | 0.1176 | 0.3003 | 0.0006 | 0.3865 | |||
| Du321 | American hybrid 3382 | 0.0002 | 0.9998 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0.828 | 0.172 | 0 | |||
| Chuan273 | (Foreign hybrid 785783 × QPM69 population)BC2 | 0.204 | 0.796 | 0.1426 | 0.5208 | 0.3366 | 0.1324 | 0.0848 | 0.5142 | 0.2686 | 0.0002 | 0.0006 | 0.5147 | 0.0394 | 0.2514 | 0.1937 | |||
| Nan381 | Ai351/B9001 // Ai351BC2 | 0.5584 | 0.4416 | 0.4927 | 0.0304 | 0.4769 | 0.4517 | 0.1901 | 0.0262 | 0.332 | 0.0974 | 0.077 | 0.0048 | 0.034 | 0.0826 | 0.7043 | |||
| 7854 | Ye478 × 156 | 0.0002 | 0.9998 | 0 | 0.4789 | 0.5211 | 0 | 0 | 0.4607 | 0.5393 | 0 | 0 | 0.4476 | 0.5524 | 0 | 0 | |||
| 18-9-101 | Nan99 / Nan70-3 Nanxiao18 | 0.9368 | 0.0632 | 0.921 | 0.051 | 0.028 | 0.921 | 0.001 | 0.04 | 0.038 | 0.8709 | 0 | 0.0364 | 0.0168 | 0.0036 | 0.0722 | |||
| ZY2247 | ZY2247 | 0.982 | 0.018 | 0.9528 | 0.0472 | 0 | 0.9566 | 0.0002 | 0.0428 | 0.0004 | 0.9546 | 0 | 0.044 | 0.0008 | 0.0004 | 0.0002 | |||
| Q78 | Q78 | 0.4234 | 0.5766 | 0.3433 | 0.2078 | 0.4489 | 0.1318 | 0.5653 | 0.2152 | 0.0876 | 0.1542 | 0.5381 | 0.2024 | 0.048 | 0.0466 | 0.0108 | |||
| Nan637 | Nan637 | 0.7518 | 0.2482 | 0.7055 | 0.0046 | 0.2899 | 0.6661 | 0.1322 | 0.0012 | 0.2004 | 0.6114 | 0.091 | 0.0016 | 0.0798 | 0.1376 | 0.0786 | |||
| Nan202 | Zong3 / Niu2-1 | 0.0382 | 0.9618 | 0.0094 | 0.836 | 0.1546 | 0.0044 | 0.0358 | 0.831 | 0.1288 | 0.0006 | 0.025 | 0.8194 | 0.0802 | 0.0018 | 0.073 | |||
| 78599-211 | Derived from Pioneer hybrid 78599 | 0.1732 | 0.8268 | 0.0002 | 0.9998 | 0 | 0.0004 | 0.0002 | 0.9994 | 0 | 0 | 0 | 0.9996 | 0 | 0.0002 | 0.0002 | |||
| YA3237 | Zheng32 × S37 | 0.659 | 0.341 | 0.5843 | 0.0024 | 0.4133 | 0.572 | 0.0978 | 0.0006 | 0.3296 | 0.5279 | 0.0484 | 0.001 | 0.1142 | 0.3061 | 0.0024 | |||
| JS0251 | Dan85566/E28 second circle line | 0.566 | 0.434 | 0.478 | 0.0008 | 0.5212 | 0.4645 | 0.109 | 0 | 0.4265 | 0.2485 | 0.0354 | 0.0002 | 0.2166 | 0.0768 | 0.4225 | |||
| W284 | W284 | 0.7372 | 0.2628 | 0.6784 | 0.004 | 0.3176 | 0.6711 | 0.0684 | 0.0006 | 0.2599 | 0.5342 | 0.0018 | 0.0014 | 0.0602 | 0.1886 | 0.2138 | |||
| Zheng28 | Zheng28 | 0.15 | 0.85 | 0.0748 | 0.0552 | 0.87 | 0.125 | 0.0012 | 0.0204 | 0.8534 | 0.134 | 0.0028 | 0.001 | 0.8614 | 0 | 0.0008 | |||
| Zheng22 | E28/Duqing second circle line | 0.026 | 0.974 | 0 | 0.0008 | 0.9992 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0.4466 | 0.5534 | 0 | |||
| Zheng99 | Zheng99 | 0.958 | 0.042 | 0.937 | 0.0618 | 0.0012 | 0.9398 | 0.0002 | 0.0568 | 0.0032 | 0.9436 | 0 | 0.036 | 0.0024 | 0.0178 | 0.0002 | |||
| Dan3130 | Derived from Pioneer hybrid 78599 | 0.2142 | 0.7858 | 0.1454 | 0.8542 | 0.0004 | 0.1198 | 0.0386 | 0.8411 | 0.0004 | 0.0942 | 0.0016 | 0.8248 | 0 | 0.0138 | 0.0656 | |||
| LC955 | LC955 | 0.9342 | 0.0658 | 0.9084 | 0.0162 | 0.0754 | 0.903 | 0.0318 | 0.0076 | 0.0576 | 0.8215 | 0.0068 | 0.005 | 0.0018 | 0.0442 | 0.1207 | |||
| Dan599 | Derived from Pioneer hybrid 78599 | 0.0932 | 0.9068 | 0.0536 | 0.8154 | 0.131 | 0.0518 | 0.0002 | 0.8066 | 0.1414 | 0.0334 | 0 | 0.8002 | 0.0914 | 0.0404 | 0.0346 | |||
| LJ-2 | American hybrid Liaojian | 0.901 | 0.099 | 0.8734 | 0.0196 | 0.107 | 0.8668 | 0.0396 | 0.009 | 0.0846 | 0.7996 | 0.0106 | 0.0086 | 0.006 | 0.0758 | 0.0994 | |||
| Liao68 | Unknown | 0.326 | 0.674 | 0.2637 | 0.5331 | 0.2032 | 0.2526 | 0.0792 | 0.5333 | 0.1348 | 0.1238 | 0 | 0.5406 | 0 | 0.2899 | 0.0456 | |||
| Liao7996 | 7922 / D9046 | 0.057 | 0.943 | 0.0002 | 0.0122 | 0.9876 | 0.0012 | 0.0002 | 0.003 | 0.9956 | 0 | 0 | 0.0008 | 0.8038 | 0.171 | 0.0244 | |||
| Liao3053 | (7922 × B68) / 5003 | 0.4704 | 0.5296 | 0.3375 | 0.0008 | 0.6617 | 0.1608 | 0.5521 | 0 | 0.2871 | 0 | 0.4351 | 0 | 0.0004 | 0.3156 | 0.2489 | |||
| Liao5144 | 7922 / 5003 | 0.1826 | 0.8174 | 0.0138 | 0.0008 | 0.9854 | 0.0622 | 0.0068 | 0 | 0.931 | 0.0328 | 0 | 0 | 0.7749 | 0.1275 | 0.0648 | |||
| Liao7890 | 7922 / 8001 | 0.0002 | 0.9998 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0.964 | 0.036 | 0 | |||
| Liao2379 | Liao2379 | 0.4502 | 0.5498 | 0.3543 | 0.0124 | 0.6333 | 0.3112 | 0.225 | 0.0064 | 0.4574 | 0.0914 | 0.1034 | 0.0118 | 0.082 | 0.3808 | 0.3307 | |||
| Wa138 | Luda Red bone germplasm | 0.4912 | 0.5088 | 0.4191 | 0.1179 | 0.4631 | 0.3386 | 0.2542 | 0.1146 | 0.2926 | 0.016 | 0.1632 | 0.0972 | 0.0492 | 0.0622 | 0.6123 | |||
| Ji1037 | Mo17 / suwan1 // MO17 | 0.54 | 0.46 | 0.4867 | 0.1476 | 0.3657 | 0.458 | 0.1272 | 0.1394 | 0.2754 | 0.1478 | 0.0484 | 0.1046 | 0.0752 | 0 | 0.6241 | |||
| TS6278 | TS6278 | 0.125 | 0.875 | 0.0126 | 0.101 | 0.8864 | 0.0484 | 0.0582 | 0.0708 | 0.8226 | 0 | 0.0464 | 0.0378 | 0.6957 | 0.0142 | 0.2059 | |||
| ZM28 | ZM28 | 0.0014 | 0.9986 | 0 | 0.9808 | 0.0192 | 0 | 0.0224 | 0.9766 | 0.001 | 0 | 0.0006 | 0.9672 | 0 | 0.032 | 0.0002 | |||
| 891 | Derived from Pioneer hybrid 78599 | 0.0012 | 0.9988 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |||
| K22 | K11 × Ye478 | 0.2262 | 0.7738 | 0.116 | 0.022 | 0.862 | 0.051 | 0.2853 | 0.014 | 0.6497 | 0.0548 | 0.2493 | 0.0002 | 0.6265 | 0.0002 | 0.069 | |||
| Dan9046 | 7922 × 5003 | 0.1088 | 0.8912 | 0.0384 | 0.4303 | 0.5313 | 0.0804 | 0.001 | 0.3964 | 0.5222 | 0.0814 | 0 | 0.3788 | 0.4802 | 0.0586 | 0.001 | |||
| PHB09 | American inbred line,Reid population | 0.3456 | 0.6544 | 0.2406 | 0.0638 | 0.6956 | 0.208 | 0.2062 | 0.0616 | 0.5242 | 0 | 0 | 0 | 0 | 1 | 0 | |||
| CN9802 | American inbred line,Reid population | 0.3562 | 0.6438 | 0.224 | 0.0016 | 0.7744 | 0.2012 | 0.186 | 0.0002 | 0.6126 | 0.0006 | 0.0642 | 0.001 | 0.1276 | 0.4915 | 0.3151 | |||
| Dan598 | Dan598 | 0.4372 | 0.5628 | 0.3883 | 0.3569 | 0.2549 | 0.3192 | 0.2072 | 0.3656 | 0.108 | 0.0912 | 0.0912 | 0.3463 | 0 | 0.0804 | 0.3909 | |||
| Shen135 | Derived from Pioneer hybrid 78599 | 0.044 | 0.956 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |||
| Shen136 | Derived from Pioneer hybrid 78599 | 0.2196 | 0.7804 | 0.151 | 0.4415 | 0.4075 | 0.1323 | 0.1191 | 0.4345 | 0.3142 | 0 | 0.0016 | 0.4313 | 0.0188 | 0.3129 | 0.2354 | |||
| Shen137 | Derived from Pioneer hybrid 6JK111 | 0.108 | 0.892 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |||
| SH15 | Derived from Pioneer hybrid 78599 | 0.1568 | 0.8432 | 0.0032 | 0.9754 | 0.0214 | 0.0016 | 0 | 0.9634 | 0.035 | 0.0002 | 0 | 0.9528 | 0.0138 | 0.0222 | 0.011 | |||
| 871 | Derived from Pioneer hybrid 87001 | 0.151 | 0.849 | 0.0956 | 0.6664 | 0.238 | 0.0228 | 0.1944 | 0.673 | 0.1098 | 0.0448 | 0.1746 | 0.6573 | 0.1228 | 0.0002 | 0.0002 | |||
| P138 | Derived from Pioneer hybrid 78599 | 0.0722 | 0.9278 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |||
| Qi319 | Derived from Pioneer hybrid 78599 | 0.0002 | 0.9998 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |||
| 178 | Derived from Pioneer hybrid 78599 | 0.045 | 0.955 | 0.0002 | 0.9998 | 0 | 0.0004 | 0 | 0.9992 | 0.0004 | 0 | 0 | 0.9862 | 0 | 0.0002 | 0.0136 | |||
| 7327 | 7327 | 0.642 | 0.358 | 0.5814 | 0.0178 | 0.4008 | 0.5167 | 0.2276 | 0.013 | 0.2426 | 0.2422 | 0.1294 | 0.0028 | 0.004 | 0.1126 | 0.509 | |||
| YS0 | YS0 | 0.5902 | 0.4098 | 0.5109 | 0.004 | 0.4851 | 0.0004 | 0.9996 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |||
| Ye478 | U8112 × Shen5003 | 0.0002 | 0.9998 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | |||
| 698-3 | Derived from Pioneer hybrid 78599 | 0.3672 | 0.6328 | 0.25 | 0.0292 | 0.7207 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |||
| JH961 | Derived from Pioneer hybrid 78599 | 0.2878 | 0.7122 | 0.2373 | 0.5614 | 0.2013 | 0.1978 | 0.125 | 0.5663 | 0.111 | 0.182 | 0.0824 | 0.5638 | 0.015 | 0.1452 | 0.0116 | |||
| JH59 | Derived from Pioneer hybrid 78599 | 0.0038 | 0.9962 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |||
| Qi205 | vAi141 × Zhongxi017/QPM70 | 0.5332 | 0.4668 | 0.4557 | 0.0384 | 0.5059 | 0.4258 | 0.1721 | 0.036 | 0.3661 | 0.1478 | 0.0458 | 0.0376 | 0.0004 | 0.3645 | 0.4039 | |||
| Dan340 | White bone Lu9 × glume maize | 0.6052 | 0.3948 | 0.5347 | 0.0016 | 0.4637 | 0.5172 | 0.1363 | 0.0006 | 0.3459 | 0 | 0 | 0 | 0 | 0.0004 | 0.9996 | |||
| CA211 | CA211 | 0.5122 | 0.4878 | 0.4524 | 0.1868 | 0.3608 | 0.3765 | 0.2322 | 0.1856 | 0.2058 | 0.1795 | 0.1805 | 0.1625 | 0.0696 | 0.0002 | 0.4078 | |||
| DYS | DYS | 0.3652 | 0.6348 | 0.2795 | 0.1318 | 0.5887 | 0.2272 | 0.2212 | 0.1296 | 0.4221 | 0 | 0.0564 | 0.1012 | 0.0004 | 0.5518 | 0.2902 | |||
| Liao6082 | 5003、7922、U8112、Zheng32、N-46、B73 integrative line | 0.3 | 0.7 | 0.0998 | 0 | 0.9002 | 0.1158 | 0.0676 | 0 | 0.8166 | 0.0863 | 0.0086 | 0 | 0.4899 | 0.3754 | 0.0398 | |||
| Lu2458 | Lu2458 | 0.194 | 0.806 | 0.1128 | 0.0526 | 0.8346 | 0.144 | 0.0002 | 0.0336 | 0.8222 | 0.0474 | 0 | 0.0168 | 0.636 | 0.0814 | 0.2183 | |||
| Ji477 | Mo17 radiation | 0.281 | 0.719 | 0.1908 | 0.027 | 0.7822 | 0.2092 | 0.0834 | 0.0084 | 0.6991 | 0.0366 | 0.0052 | 0.0206 | 0.3136 | 0.3184 | 0.3056 | |||
| Zheng29 | Zheng29 | 0.1862 | 0.8138 | 0.0354 | 0.0042 | 0.9604 | 0.0606 | 0.0672 | 0 | 0.8722 | 0.012 | 0.0132 | 0.0006 | 0.5864 | 0.3828 | 0.005 | |||
| ZH64 | Derived from Pioneer hybrid 64 | 0.486 | 0.514 | 0.4219 | 0.1638 | 0.4143 | 0.3703 | 0.1893 | 0.1599 | 0.2805 | 0.1414 | 0.1148 | 0.1436 | 0.0876 | 0.0526 | 0.46 | |||
| H921 | 78599 × (6166 + 6066 + 6165) | 0.3592 | 0.6408 | 0.2914 | 0.2948 | 0.4138 | 0.2111 | 0.2643 | 0.3053 | 0.2193 | 0.0534 | 0.1598 | 0.2985 | 0.0026 | 0.2392 | 0.2464 | |||
| 1572 | 5003 × Luda Red bone line | 0.339 | 0.661 | 0.2629 | 0.088 | 0.6491 | 0.3035 | 0.0178 | 0.065 | 0.6137 | 0.0796 | 0 | 0.011 | 0.4656 | 0 | 0.4438 | |||
| Ji992 | Ji992 | 0.239 | 0.761 | 0.1581 | 0.1395 | 0.7025 | 0.198 | 0.0014 | 0.1124 | 0.6881 | 0 | 0 | 0.0642 | 0.4242 | 0.0008 | 0.5108 | |||
| 9614 | Tropical germplasm | 0.296 | 0.704 | 0.2121 | 0.1082 | 0.6797 | 0.2264 | 0.0736 | 0.0982 | 0.6018 | 0.0828 | 0.0434 | 0.064 | 0.4648 | 0.0002 | 0.3447 | |||
| 434 | 434 | 0.3692 | 0.6308 | 0.2918 | 0.156 | 0.5522 | 0.3254 | 0.016 | 0.1414 | 0.5172 | 0.086 | 0 | 0.1303 | 0.2161 | 0.1839 | 0.3838 | |||
| H10 | Tropical germplasm | 0.4232 | 0.5768 | 0.3513 | 0.1947 | 0.454 | 0.3276 | 0.1332 | 0.1898 | 0.3494 | 0.2004 | 0.0786 | 0.1662 | 0.2354 | 0.0408 | 0.2787 | |||
| 4379 | 4379 | 0.604 | 0.396 | 0.5521 | 0.2118 | 0.236 | 0.5458 | 0.06 | 0.2048 | 0.1894 | 0.3437 | 0.0006 | 0.1964 | 0.0018 | 0.1486 | 0.3089 | |||
| 4011 | Derived from landrace | 0.6402 | 0.3598 | 0.5761 | 0.0026 | 0.4213 | 0.5749 | 0.0792 | 0.0006 | 0.3453 | 0.1694 | 0 | 0.0008 | 0.0018 | 0.1278 | 0.7003 | |||
| HuangC | (Huangxiao162 × Zi330 / 02) × Tuxpeno | 0.37 | 0.63 | 0.2895 | 0.109 | 0.6015 | 0.3311 | 0.004 | 0.0828 | 0.5821 | 0.1952 | 0.0002 | 0.0562 | 0.4627 | 0.0002 | 0.2855 | |||
| Tie7922 | American hybrid 3382 | 0.038 | 0.962 | 0.0002 | 0.0466 | 0.9532 | 0.0016 | 0.0114 | 0.0386 | 0.9484 | 0.03 | 0.002 | 0.0238 | 0.9366 | 0.0002 | 0.0074 | |||
| 9HT1804 | 9HT1804 | 0.4378 | 0.5622 | 0.3735 | 0.2478 | 0.3787 | 0.3433 | 0.1358 | 0.2456 | 0.2753 | 0.2272 | 0.0858 | 0.2264 | 0.1804 | 0.023 | 0.2572 | |||
| M165 | M165 | 0.3492 | 0.6508 | 0.2667 | 0.0588 | 0.6745 | 0.2284 | 0.2074 | 0.0602 | 0.5039 | 0.1311 | 0.1527 | 0.043 | 0.3609 | 0.0906 | 0.2217 | |||
| H21 | Huangzaosi × H84 | 0.968 | 0.032 | 0.9416 | 0.0584 | 0 | 0.945 | 0.0002 | 0.0544 | 0.0004 | 0.9558 | 0 | 0.0416 | 0.0004 | 0.0022 | 0 | |||
| 4866 | Ye478 improved line | 0.217 | 0.783 | 0.1196 | 0.0428 | 0.8375 | 0.1782 | 0.0054 | 0.0022 | 0.8142 | 0.1163 | 0.0018 | 0.0002 | 0.7306 | 0.0018 | 0.1493 | |||
| 07G83 | 07G83 | 0.488 | 0.512 | 0.4219 | 0.2098 | 0.3683 | 0.3531 | 0.2326 | 0.2196 | 0.1948 | 0.1712 | 0.1288 | 0.2056 | 0.0004 | 0.219 | 0.2749 | |||
| LJS-1 | LJS-1 | 0.4112 | 0.5888 | 0.3583 | 0.3241 | 0.3175 | 0.1724 | 0.4493 | 0.3433 | 0.035 | 0.1962 | 0.4188 | 0.3352 | 0.0254 | 0.0238 | 0.0006 | |||
| Yi99-19 | Zhengda hybrid 99-19 | 0.853 | 0.147 | 0.816 | 0.0146 | 0.1694 | 0.783 | 0.103 | 0.0086 | 0.1054 | 0.7326 | 0.0546 | 0.0106 | 0.0022 | 0.1826 | 0.0174 | |||
| LZM004 | Tropical germplasm | 0.1844 | 0.8156 | 0.062 | 0.938 | 0 | 0.0676 | 0 | 0.9324 | 0 | 0.0728 | 0 | 0.9268 | 0 | 0.0002 | 0.0002 | |||
| LZM05-1-1 | Tropical germplasm | 0.9992 | 0.0008 | 0.9976 | 0.0024 | 0 | 0.9998 | 0.0002 | 0 | 0 | 0.9984 | 0 | 0.0004 | 0.0004 | 0.0002 | 0.0006 | |||
| LZM6-1-1 | Tropical germplasm | 0.9948 | 0.0052 | 0.9678 | 0.0316 | 0.0006 | 0.9412 | 0.0424 | 0.016 | 0.0004 | 0.916 | 0.0452 | 0.0164 | 0.0004 | 0.0002 | 0.0218 | |||
| LZM025 | Tropical germplasm | 0.269 | 0.731 | 0.2176 | 0.6302 | 0.1522 | 0.1378 | 0.2106 | 0.64 | 0.0116 | 0.1489 | 0.1833 | 0.627 | 0.0338 | 0.0028 | 0.0042 | |||
| W8071 | Tropical germplasm | 0.8448 | 0.1552 | 0.8064 | 0.0136 | 0.18 | 0.7983 | 0.0524 | 0.0046 | 0.1447 | 0.5919 | 0.0006 | 0.0012 | 0.0004 | 0.0786 | 0.3273 | |||
| M11 | Derived from Mobai 1 | 0.9114 | 0.0886 | 0.8856 | 0.01 | 0.1044 | 0.8842 | 0.0192 | 0.0018 | 0.0948 | 0.8126 | 0.0028 | 0.0016 | 0.0538 | 0.0058 | 0.1234 | |||
| 1221CL8 | 1221CL8 | 0.823 | 0.177 | 0.7846 | 0.0092 | 0.2062 | 0.7877 | 0.0246 | 0.002 | 0.1857 | 0.6251 | 0.0006 | 0.0018 | 0.0414 | 0.0876 | 0.2436 | |||
| QBII-1 | 1221CL13 | 0.7814 | 0.2186 | 0.7359 | 0.0108 | 0.2533 | 0.7039 | 0.1228 | 0.0038 | 0.1696 | 0.4944 | 0.0566 | 0.0038 | 0.0068 | 0.0728 | 0.3656 | |||
| 1221CL11 | 1221CL11 | 0.9124 | 0.0876 | 0.8872 | 0.0118 | 0.101 | 0.8884 | 0.0146 | 0.002 | 0.095 | 0.8193 | 0.0014 | 0.0018 | 0.0554 | 0.005 | 0.1171 | |||
| ML1108 | ML1108 | 0.4192 | 0.5808 | 0.3176 | 0.0706 | 0.6118 | 0.2153 | 0.3301 | 0.0732 | 0.3814 | 0.0334 | 0.2102 | 0.0698 | 0.066 | 0.3985 | 0.222 | |||
| ML1120 | ML1120 | 0.5752 | 0.4248 | 0.4981 | 0.0252 | 0.4767 | 0.0004 | 0.9782 | 0.0214 | 0 | 0 | 0.9998 | 0 | 0 | 0 | 0.0002 | |||
| W7475 | 12127475 | 0.1494 | 0.8506 | 0.0892 | 0.5299 | 0.3809 | 0.0154 | 0.2101 | 0.5368 | 0.2377 | 0 | 0.1527 | 0.5062 | 0.1671 | 0.0308 | 0.1433 | |||
| Liao147-8 | Liao147-822411 | 0.465 | 0.535 | 0.3897 | 0.1626 | 0.4477 | 0.0912 | 0.7233 | 0.1851 | 0.0004 | 0.103 | 0.7013 | 0.1758 | 0.0004 | 0.0194 | 0.0002 | |||
| PN0504-8 | Derived from Pioneer hybrid PN0504 | 0.271 | 0.729 | 0.1965 | 0.3151 | 0.4884 | 0.1712 | 0.1464 | 0.3153 | 0.3671 | 0.0616 | 0.0608 | 0.3151 | 0.1318 | 0.2673 | 0.1634 | |||
| KS001 | Derived from American hybrid X1141P | 0.4472 | 0.5528 | 0.3375 | 0.0024 | 0.6601 | 0.2707 | 0.2803 | 0.0006 | 0.4485 | 0 | 0.0869 | 0.0002 | 0 | 0.6188 | 0.2942 | |||
| A318 | S37 space induced mutation | 0.951 | 0.049 | 0.929 | 0.0576 | 0.0134 | 0.8966 | 0.0544 | 0.0482 | 0.0008 | 0.8444 | 0.024 | 0.0358 | 0.0004 | 0.0002 | 0.0952 | |||
| 5220-2 | Unknown | 0.1762 | 0.8238 | 0.0834 | 0.1764 | 0.7402 | 0.0908 | 0.084 | 0.1709 | 0.6543 | 0.0008 | 0.0488 | 0.1592 | 0.4571 | 0.111 | 0.223 | |||
| SAM1001 | Tropical germplasm TI5604 | 0.85 | 0.15 | 0.822 | 0.0546 | 0.1234 | 0.8198 | 0.023 | 0.044 | 0.1132 | 0.7145 | 0.0018 | 0.033 | 0.0614 | 0.0002 | 0.189 | |||
| SAM3001 | Unknown hybrid 3001 | 0.2264 | 0.7736 | 0.181 | 0.6742 | 0.1448 | 0.1481 | 0.1054 | 0.6719 | 0.0746 | 0 | 0.0166 | 0.6549 | 0 | 0.044 | 0.2845 | |||
| 975-12 | 698-3 / unknown inbred line | 0.113 | 0.887 | 0.0014 | 0.0877 | 0.9109 | 0 | 0.5824 | 0 | 0.4176 | 0 | 0.5739 | 0 | 0.4261 | 0 | 0 | |||
| 18-599 | Derived from Pioneer hybrid 78599 | 0.0168 | 0.9832 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |||
| SCML202 | (48-2 × 5003)space induced mutation × foreign hybrid | 0.0348 | 0.9652 | 0.0008 | 0.5614 | 0.4378 | 0.0016 | 0.0372 | 0.563 | 0.3982 | 0.0006 | 0.034 | 0.5718 | 0.2279 | 0.1521 | 0.0136 | |||
| SCML1950 | American hybrid 441950 | 0.3688 | 0.6312 | 0.263 | 0.0106 | 0.7264 | 0.2821 | 0.0506 | 0.0006 | 0.6667 | 0.1058 | 0.001 | 0.0002 | 0.4658 | 0.0712 | 0.3559 | |||
| 08-641 | Derived from Pioneer hybrid Y78641 | 0.5462 | 0.4538 | 0.4747 | 0.1044 | 0.4209 | 0.2912 | 0.4728 | 0.117 | 0.119 | 0.1062 | 0.3761 | 0.0934 | 0.0102 | 0.0018 | 0.4123 | |||
| 48-2 | (Mo17 / Dan340)// 6 unknown lines | 0.85 | 0.15 | 0.7795 | 0.0008 | 0.2197 | 0.7331 | 0.1556 | 0 | 0.1112 | 0.4669 | 0.0594 | 0 | 0 | 0.0006 | 0.4731 | |||
| SCML103 | 698-3 / unknown line | 0.1584 | 0.8416 | 0.0184 | 0.0228 | 0.9588 | 0 | 0.5184 | 0 | 0.4816 | 0 | 0.5366 | 0 | 0.4632 | 0.0002 | 0 | |||
| ES40 | Landrace Linshuidadudu | 0.6652 | 0.3348 | 0.6102 | 0.0456 | 0.3442 | 0.5906 | 0.12 | 0.0384 | 0.251 | 0.3481 | 0.043 | 0.0194 | 0.045 | 0.039 | 0.5054 | |||
| SCML203 | (48-2 × 5003)space induced mutation × foreign hybrid | 0.0052 | 0.9948 | 0.0002 | 0.4223 | 0.5775 | 0.0012 | 0.0032 | 0.4276 | 0.568 | 0 | 0.0002 | 0.424 | 0.3455 | 0.1745 | 0.0558 | |||
| LH8012 | Yunrui9 (YML46 / YML8711) second circle line | 0.2742 | 0.7258 | 0.1978 | 0.253 | 0.5492 | 0.1178 | 0.2525 | 0.2459 | 0.3838 | 0.0008 | 0.2198 | 0.231 | 0.2663 | 0.0002 | 0.2819 | |||
| 9LB050 | 9LB050 | 0.9098 | 0.0902 | 0.8822 | 0.0362 | 0.0816 | 0.8753 | 0.033 | 0.0288 | 0.063 | 0.7782 | 0.002 | 0.0194 | 0.0044 | 0.0058 | 0.1902 | |||
| TM012 | TM012 | 0.244 | 0.756 | 0.1618 | 0.3566 | 0.4816 | 0.1284 | 0.1576 | 0.3556 | 0.3584 | 0.0012 | 0.0918 | 0.3511 | 0.1524 | 0.1518 | 0.2517 | |||
| 1323 | Hybrid 1323 | 0.463 | 0.537 | 0.4208 | 0.398 | 0.1812 | 0.381 | 0.1244 | 0.3926 | 0.102 | 0.2627 | 0.0706 | 0.383 | 0.006 | 0.0516 | 0.2261 | |||
| 975-13 | 698-3 / unknown line | 0.1732 | 0.8268 | 0.0318 | 0.006 | 0.9622 | 0 | 0.6494 | 0 | 0.3506 | 0 | 0.6713 | 0 | 0.3147 | 0.014 | 0 | |||
| YZ15BC-2 | YZ15BC-2 | 0.2952 | 0.7048 | 0.2217 | 0.3013 | 0.477 | 0.1728 | 0.1912 | 0.3003 | 0.3357 | 0.001 | 0.1307 | 0.2898 | 0.1227 | 0.0977 | 0.3582 | |||
| SCML2054 | Hybrid 2054 | 0.1532 | 0.8468 | 0.0632 | 0.2252 | 0.7115 | 0.0804 | 0.0568 | 0.2162 | 0.6465 | 0.0018 | 0.0256 | 0.2053 | 0.4718 | 0.0812 | 0.2143 | |||
| 10WRB115 | 10WRB115 | 0.1514 | 0.8486 | 0.0946 | 0.5419 | 0.3635 | 0.024 | 0.2022 | 0.5481 | 0.2256 | 0 | 0.1473 | 0.5202 | 0.1557 | 0.0294 | 0.1475 | |||
| 10WRA120 | 10WRA120 | 0.2676 | 0.7324 | 0.1565 | 0.0456 | 0.798 | 0.0968 | 0.2704 | 0.0444 | 0.5884 | 0 | 0.1958 | 0.0012 | 0.3942 | 0.039 | 0.3698 | |||
| 08WSC51 | 08WSC51-221 | 0.2486 | 0.7514 | 0.1462 | 0.0894 | 0.7644 | 0.118 | 0.186 | 0.0802 | 0.6157 | 0.0044 | 0.1284 | 0.0792 | 0.3725 | 0.2306 | 0.185 | |||
| 08WSC179 | 08WSC179-31 | 0.7782 | 0.2218 | 0.7227 | 0.004 | 0.2733 | 0.4227 | 0.5771 | 0.0002 | 0 | 0.3942 | 0.5596 | 0.0002 | 0 | 0.0002 | 0.0458 | |||
| 08WSC187 | 08WSC187-211 | 0.1912 | 0.8088 | 0.122 | 0.4991 | 0.3789 | 0.0004 | 0.44 | 0.485 | 0.0746 | 0.0006 | 0.4384 | 0.471 | 0.0874 | 0.0026 | 0 | |||
| 08WSC200 | 08WSC200-11 | 0.0892 | 0.9108 | 0.053 | 0.7884 | 0.1586 | 0.0538 | 0.0192 | 0.7859 | 0.1411 | 0.0724 | 0.004 | 0.7736 | 0.1066 | 0.0394 | 0.004 | |||
| 08WSC204 | 08WSC204-21 | 0.272 | 0.728 | 0.2194 | 0.5982 | 0.1824 | 0.2177 | 0.044 | 0.592 | 0.1463 | 0.0744 | 0 | 0.5871 | 0.0386 | 0.1116 | 0.1884 | |||
| 08WSC237 | 08WSC237-111 | 0.3082 | 0.6918 | 0.2234 | 0.256 | 0.5206 | 0.0824 | 0.4398 | 0.2539 | 0.2239 | 0.0102 | 0.3964 | 0.249 | 0.0842 | 0.16 | 0.1002 | |||
| 08WSC257 | 08WSC257-122 | 0.4292 | 0.5708 | 0.3699 | 0.3097 | 0.3203 | 0.2167 | 0.409 | 0.3213 | 0.053 | 0.2308 | 0.3894 | 0.3022 | 0.0718 | 0.0048 | 0.001 | |||
| CIMMYT-2 | CIMMYT-2 | 0.9974 | 0.0026 | 0.9782 | 0.0212 | 0.0006 | 0.9802 | 0.0068 | 0.0126 | 0.0004 | 0.9806 | 0.0032 | 0.0134 | 0.0008 | 0.001 | 0.001 | |||
| CIMMYT-1 | CIMMYT-1 | 0.9724 | 0.0276 | 0.9624 | 0.0198 | 0.0178 | 0.9542 | 0.0202 | 0.0086 | 0.017 | 0.941 | 0.0078 | 0.0042 | 0.0296 | 0.0002 | 0.0172 | |||
| GP30-1 | Unknown | 0.2938 | 0.7062 | 0.2136 | 0.2362 | 0.5502 | 0.2068 | 0.0966 | 0.2198 | 0.4768 | 0.0088 | 0.0502 | 0.1919 | 0.3237 | 0.0002 | 0.4252 | |||
| GP66-1 | Unknown | 0.447 | 0.553 | 0.3926 | 0.3446 | 0.2628 | 0.3065 | 0.2422 | 0.3463 | 0.105 | 0.1714 | 0.1838 | 0.3318 | 0.0098 | 0.023 | 0.2802 | |||
| S37 | Derived from suwan1 | 0.9936 | 0.0064 | 0.9784 | 0.0184 | 0.0032 | 0.935 | 0.0632 | 0.0014 | 0.0004 | 0.918 | 0.0578 | 0.0014 | 0.0004 | 0.0002 | 0.0222 | |||
| T32 | Derived from suwan germplasm | 0.9992 | 0.0008 | 0.9992 | 0.0008 | 0 | 0.9998 | 0.0002 | 0 | 0 | 0.9998 | 0 | 0.0002 | 0 | 0 | 0 | |||
| M3-0 | Tropical germplasm | 0.864 | 0.136 | 0.8295 | 0.0869 | 0.0836 | 0.8074 | 0.0678 | 0.0858 | 0.039 | 0.7594 | 0.034 | 0.0776 | 0.0004 | 0.071 | 0.0576 | |||
| BJ005 | BJ005 | 0.164 | 0.836 | 0.0666 | 0.106 | 0.8274 | 0.1012 | 0.041 | 0.0732 | 0.7846 | 0.0012 | 0.0128 | 0.0584 | 0.5821 | 0.086 | 0.2595 | |||
| 2082-11 | 2082-11 | 0.3532 | 0.6468 | 0.2682 | 0.2136 | 0.5182 | 0.0022 | 0.7724 | 0.2226 | 0.0028 | 0.0006 | 0.7251 | 0.1927 | 0 | 0.0812 | 0.0004 | |||
| GD003 | 11GD003 | 0.6164 | 0.3836 | 0.5496 | 0.0382 | 0.4122 | 0.4405 | 0.318 | 0.0422 | 0.1993 | 0.4273 | 0.2953 | 0.0256 | 0.1864 | 0.001 | 0.0644 | |||
| 03FLUSA10 | American hybrid 03FLUSA10 | 0.5802 | 0.4198 | 0.528 | 0.1812 | 0.2908 | 0.4441 | 0.2353 | 0.1869 | 0.1337 | 0.3417 | 0.1828 | 0.1832 | 0.0048 | 0.1272 | 0.1604 | |||
| XS021 | XS021 | 0.4834 | 0.5166 | 0.4274 | 0.1628 | 0.4098 | 0.4196 | 0.0924 | 0.1549 | 0.3331 | 0.1758 | 0.0412 | 0.1406 | 0.1128 | 0.057 | 0.4727 | |||
| 9HT1736 | Unknown | 0.966 | 0.034 | 0.947 | 0.0318 | 0.0212 | 0.932 | 0.0398 | 0.0248 | 0.0034 | 0.904 | 0.0178 | 0.018 | 0.0014 | 0.011 | 0.0478 | |||
| 81565 | TuxpenoPBC15S1 ×(JinO3 × Huobai) | 0.8492 | 0.1508 | 0.8091 | 0.0134 | 0.1775 | 0.8132 | 0.028 | 0.0046 | 0.1542 | 0.6788 | 0.0006 | 0.0026 | 0.0286 | 0.0853 | 0.2041 | |||
| ZD0502-23111 | Hybrid ZD0502 | 0.656 | 0.344 | 0.6295 | 0.3397 | 0.0308 | 0.6331 | 0.001 | 0.3335 | 0.0324 | 0.5641 | 0 | 0.3246 | 0.0004 | 0.0174 | 0.0935 | |||
| 09YT20919 | Tropical integrative line AP5 | 0.9992 | 0.0008 | 0.9968 | 0.0032 | 0 | 0.9986 | 0.0002 | 0.0006 | 0.0006 | 0.9938 | 0 | 0.001 | 0.0008 | 0.002 | 0.0024 | |||
| LH7556 | LH7556 | 0.0004 | 0.9996 | 0 | 0.502 | 0.498 | 0.0004 | 0.0002 | 0.4978 | 0.5016 | 0 | 0.0004 | 0.4922 | 0.4538 | 0.0016 | 0.052 | |||
| Mian04185-4 | Mian04185-43122 | 0.534 | 0.466 | 0.4805 | 0.152 | 0.3675 | 0.4639 | 0.1035 | 0.1449 | 0.2878 | 0.1933 | 0.0412 | 0.1291 | 0.0806 | 0.0284 | 0.5274 | |||
| Mian04185-8 | Mian04185-8411 | 0.586 | 0.414 | 0.5333 | 0.1004 | 0.3663 | 0.5157 | 0.1105 | 0.0955 | 0.2784 | 0.1931 | 0.0152 | 0.0782 | 0.0304 | 0.0628 | 0.6202 | |||
| 07WSWD146 | 07WSWD146 | 0.6538 | 0.3462 | 0.5985 | 0.0656 | 0.3359 | 0.5736 | 0.1262 | 0.062 | 0.2381 | 0.3355 | 0.0282 | 0.0568 | 0.0022 | 0.1601 | 0.4172 | |||
| SCML2031 | TF2031-2221 | 0.42 | 0.58 | 0.376 | 0.5145 | 0.1095 | 0.3587 | 0.0696 | 0.5129 | 0.0588 | 0.23 | 0.0028 | 0.5004 | 0.0004 | 0.0474 | 0.219 | |||
| TY30331-3 | TY30331-35121 | 0.648 | 0.352 | 0.5966 | 0.1096 | 0.2938 | 0.5442 | 0.1726 | 0.109 | 0.1742 | 0.4009 | 0.116 | 0.108 | 0.004 | 0.149 | 0.222 | |||
| TY30331-2 | TY30331-25111 | 0.663 | 0.337 | 0.6175 | 0.1476 | 0.235 | 0.5097 | 0.2793 | 0.1492 | 0.0618 | 0.4735 | 0.2532 | 0.1409 | 0.0344 | 0.0118 | 0.0863 | |||
| QA | 178/9782 | 0.0432 | 0.9568 | 0.0006 | 0.4342 | 0.5652 | 0.0118 | 0.0054 | 0.4306 | 0.5522 | 0 | 0 | 0.3962 | 0.415 | 0.0002 | 0.1887 | |||
| KS003 | Derived from integrative line | 0.561 | 0.439 | 0.4608 | 0.0008 | 0.5384 | 0.3016 | 0.4598 | 0.0006 | 0.238 | 0.1599 | 0.4044 | 0.0002 | 0.0698 | 0.1012 | 0.2645 | |||
| HL5049 | HL5049 | 0.151 | 0.849 | 0.1056 | 0.6973 | 0.1972 | 0.0216 | 0.2162 | 0.7085 | 0.0538 | 0.034 | 0.2003 | 0.6955 | 0.0628 | 0.0002 | 0.0072 | |||
| HL5054 | HL5054 | 0.1322 | 0.8678 | 0.0816 | 0.7012 | 0.2172 | 0.002 | 0.2649 | 0.7043 | 0.0288 | 0.0024 | 0.2507 | 0.6875 | 0.0588 | 0.0002 | 0.0004 | |||
| LLF-08 | 08-641 backcross improved line | 0.5462 | 0.4538 | 0.4748 | 0.1034 | 0.4218 | 0.2915 | 0.4729 | 0.116 | 0.1196 | 0.106 | 0.3762 | 0.092 | 0.011 | 0.002 | 0.4128 | |||
| ZD808-1 | Hybrid ZD808 | 0.5762 | 0.4238 | 0.4853 | 0.0022 | 0.5125 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |||
| 510317 | Hybrid 510317 | 0.103 | 0.897 | 0.0294 | 0.5849 | 0.3857 | 0.0002 | 0.339 | 0.5494 | 0.1114 | 0 | 0.3369 | 0.5311 | 0.1316 | 0.0002 | 0.0002 | |||
| Dan4245 | Liao7980 / Dan598 | 0.3692 | 0.6308 | 0.2974 | 0.2398 | 0.4628 | 0.2384 | 0.2334 | 0.2494 | 0.2789 | 0.0006 | 0.0814 | 0.2414 | 0.0004 | 0.3457 | 0.3305 | |||
| PZ1010-2 | PZ1010-2 | 0.5762 | 0.4238 | 0.4932 | 0.0038 | 0.503 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |||
| CMY093288 | CMY093288 | 0.4632 | 0.5368 | 0.3966 | 0.1906 | 0.4128 | 0.3087 | 0.2801 | 0.2008 | 0.2104 | 0.0272 | 0.1664 | 0.1834 | 0.0004 | 0.1492 | 0.4733 | |||
| JD7275 | Hybrid JD7275 | 0.7844 | 0.2156 | 0.75 | 0.171 | 0.079 | 0.7406 | 0.0388 | 0.1658 | 0.0548 | 0.7025 | 0.0018 | 0.1622 | 0.0052 | 0.0692 | 0.0592 | |||
| JY01-3 | Hybrid JY01 | 0.096 | 0.904 | 0.0302 | 0.5274 | 0.4424 | 0.0168 | 0.095 | 0.5202 | 0.368 | 0 | 0.0584 | 0.4884 | 0.2729 | 0.0002 | 0.1801 | |||
| Y0827 | SN203 / SN204 / SC9911 / DK656 / CS9906 integrative line | 0.4034 | 0.5966 | 0.2982 | 0.0032 | 0.6986 | 0.2938 | 0.1302 | 0.0006 | 0.5754 | 0.0602 | 0.004 | 0.01 | 0.0336 | 0.5983 | 0.2939 | |||
| Y0921 | Ye478 / 5003 / 7922 / Du32 / Cheng687 / 698-3 integrative line | 0.223 | 0.777 | 0.109 | 0.0994 | 0.7917 | 0.0022 | 0.4171 | 0.0906 | 0.4901 | 0.0032 | 0.3992 | 0.0786 | 0.3792 | 0.1395 | 0.0002 | |||
| Y1015 | Tropical integrative line AP5 | 0.9982 | 0.0018 | 0.9936 | 0.006 | 0.0004 | 0.9864 | 0.0112 | 0.002 | 0.0004 | 0.9576 | 0.0022 | 0.0012 | 0.0004 | 0.0002 | 0.0384 | |||
| Y1018 | Tropical integrative line AP5 | 0.9888 | 0.0112 | 0.9766 | 0.0216 | 0.0018 | 0.977 | 0.0004 | 0.0046 | 0.018 | 0.9594 | 0 | 0.0022 | 0.0154 | 0.0016 | 0.0214 | |||
| Y1021 | Ye478 / 5003 / 7922 / Du32 / Cheng687 / 698-3 integrative line | 0.141 | 0.859 | 0.0328 | 0.2446 | 0.7226 | 0.0004 | 0.3395 | 0.2103 | 0.4498 | 0 | 0.3634 | 0.2048 | 0.4292 | 0.0024 | 0.0002 | |||
| Y1022 | Ye478 / 5003 / 7922 / Du32 / Cheng687 / 698-3 integrative line | 0.5414 | 0.4586 | 0.4816 | 0.1184 | 0.4 | 0.3203 | 0.4269 | 0.1252 | 0.1276 | 0.2413 | 0.366 | 0.1145 | 0.028 | 0.0903 | 0.1599 | |||
| Y1032W | 273 / CH01-1112 / 634-1112 / 698-3 / Cheng687 / 7932-1211 / 5003 / 18-599 / Hai92-1 integrative line | 0.152 | 0.848 | 0.0818 | 0.4113 | 0.5069 | 0.0172 | 0.2494 | 0.4168 | 0.3166 | 0.0006 | 0.2122 | 0.4112 | 0.1488 | 0.1474 | 0.0798 | |||
| Y1032R | 273 / CH01-1112 / 634-1112 / 698-3 / Cheng687/7932-1211 / 5003 / 18-599 / Hai92-1 integrative line | 0.1482 | 0.8518 | 0.0784 | 0.4113 | 0.5103 | 0.0172 | 0.2424 | 0.4169 | 0.3235 | 0.0004 | 0.2012 | 0.41 | 0.1452 | 0.153 | 0.0902 | |||
| Y1035 | 08-641 natural abnormal plant | 0.0712 | 0.9288 | 0.002 | 0.4138 | 0.5842 | 0 | 0.4876 | 0.3594 | 0.153 | 0 | 0.4802 | 0.3627 | 0.0646 | 0.0924 | 0 | |||
| Y1038 | 08-641 natural abnormal plant | 0.2252 | 0.7748 | 0.1542 | 0.3392 | 0.5066 | 0.08 | 0.2736 | 0.3432 | 0.3032 | 0.0022 | 0.205 | 0.3466 | 0.0856 | 0.2242 | 0.1364 | |||
| Y1111 | Y1111 | 0.4182 | 0.5818 | 0.3533 | 0.1928 | 0.4539 | 0.2632 | 0.2728 | 0.1882 | 0.2758 | 0.1583 | 0.2339 | 0.1765 | 0.1351 | 0.1132 | 0.1831 | |||
| Y1127 | Y1127 | 0.9818 | 0.0182 | 0.9686 | 0.0202 | 0.0112 | 0.9666 | 0.0002 | 0.0016 | 0.0316 | 0.9546 | 0.0004 | 0.0012 | 0.03 | 0.0014 | 0.0124 | |||
| Y1027 | Derived from integrative line W1030 | 0.4072 | 0.5928 | 0.3126 | 0.0556 | 0.6318 | 0.2526 | 0.2642 | 0.0538 | 0.4294 | 0.0812 | 0.1985 | 0.0402 | 0.2488 | 0.0672 | 0.364 | |||
| L31 | 273 / CH01-1112 / 634-1112 / 698-3 / Cheng687 / 7932-1211 / 5003 / 18-599 / Hai92-1 integrative line | 0.2002 | 0.7998 | 0.1214 | 0.4061 | 0.4725 | 0.0044 | 0.3971 | 0.4103 | 0.1882 | 0 | 0.3233 | 0.3923 | 0.0306 | 0.15 | 0.1038 | |||
| L27 | 273 / CH01-1112 / 634-1112 / 698-3 / Cheng687 / 7932-1211 / 5003 / 18-599 / Hai92-1 integrative line | 0.2244 | 0.7756 | 0.153 | 0.4752 | 0.3718 | 0.0338 | 0.3534 | 0.4831 | 0.1297 | 0 | 0.2949 | 0.466 | 0.0114 | 0.1084 | 0.1192 | |||
| Y1005 | Tropical integrative line AP5 | 0.9984 | 0.0016 | 0.9962 | 0.0024 | 0.0014 | 0.949 | 0.0506 | 0 | 0.0004 | 0.8886 | 0.044 | 0.0002 | 0.0004 | 0.0002 | 0.0666 | |||
| Y0826 | Tropical integrative line AP5 | 0.9992 | 0.0008 | 0.9974 | 0.0024 | 0.0002 | 0.999 | 0.0002 | 0.0002 | 0.0006 | 0.9928 | 0 | 0.0004 | 0.0014 | 0.0022 | 0.0032 | |||
| 77 | 77 | 0.3996 | 0.6004 | 0.3245 | 0.1466 | 0.5289 | 0.3551 | 0.0186 | 0.1367 | 0.4896 | 0.1144 | 0 | 0.1232 | 0.1815 | 0.1767 | 0.4042 | |||
| BML1256 | BML1256 | 0.991 | 0.009 | 0.9592 | 0.0016 | 0.0392 | 0.9422 | 0.0436 | 0.0006 | 0.0136 | 0.8862 | 0.0226 | 0.0002 | 0.0018 | 0.0046 | 0.0846 | |||
| P801 | P801 | 0.4232 | 0.5768 | 0.359 | 0.2415 | 0.3996 | 0.2905 | 0.2334 | 0.2509 | 0.2252 | 0.0873 | 0.1323 | 0.2418 | 0.0034 | 0.1783 | 0.3569 | |||
| 10WRC64 | 10WRC64 | 0.0034 | 0.9966 | 0.0002 | 0.9326 | 0.0672 | 0.0002 | 0.0002 | 0.9322 | 0.0674 | 0 | 0 | 0.9226 | 0.0194 | 0.0002 | 0.0578 | |||
| TLL-1 | Derived from Thailand landrace | 0.9966 | 0.0034 | 0.982 | 0.0164 | 0.0016 | 0.9744 | 0.0182 | 0.0054 | 0.002 | 0.941 | 0.0044 | 0.0034 | 0.0006 | 0.0106 | 0.04 | |||
| PI43W | PI43W | 0.743 | 0.257 | 0.683 | 0.0016 | 0.3154 | 0.6556 | 0.1212 | 0.0006 | 0.2226 | 0.3453 | 0.008 | 0.0002 | 0.0004 | 0.1016 | 0.5444 | |||
| W8199 | 11278199 | 0.508 | 0.492 | 0.448 | 0.2182 | 0.3338 | 0.3307 | 0.3231 | 0.2222 | 0.124 | 0.2413 | 0.2764 | 0.2171 | 0.0044 | 0.1099 | 0.1509 | |||
| Y1114 | Tropical integrative line AP5 | 0.9992 | 0.0008 | 0.9992 | 0.0008 | 0 | 0.9996 | 0.0004 | 0 | 0 | 0.9998 | 0 | 0.0002 | 0 | 0 | 0 | |||
| LX312 | LX312 | 0.4332 | 0.5668 | 0.3491 | 0.1276 | 0.5233 | 0.0082 | 0.8488 | 0.1426 | 0.0004 | 0.0012 | 0.7975 | 0.1236 | 0 | 0.0014 | 0.0762 | |||
| SAM31152A | SAM3001 × 1152A-111 | 0.2492 | 0.7508 | 0.183 | 0.41 | 0.407 | 0.1314 | 0.2088 | 0.4139 | 0.246 | 0 | 0.0036 | 0.3839 | 0 | 0.3929 | 0.2196 | |||
| DH3732 | Denghai hybrid DH3732 | 0.2354 | 0.7646 | 0.1776 | 0.6371 | 0.1854 | 0.182 | 0.0294 | 0.6356 | 0.153 | 0.1074 | 0.0018 | 0.6275 | 0.0658 | 0.0332 | 0.1642 | |||
| BML1275 | BML1275 | 0.746 | 0.254 | 0.7046 | 0.0834 | 0.212 | 0.66 | 0.1404 | 0.0816 | 0.118 | 0.4277 | 0.0484 | 0.0552 | 0 | 0.0766 | 0.3921 | |||
| XBY13563 | XBY13563-283 | 0.827 | 0.173 | 0.791 | 0.137 | 0.072 | 0.7544 | 0.097 | 0.1368 | 0.0118 | 0.746 | 0.077 | 0.1258 | 0.0034 | 0.0474 | 0.0004 | |||
| BML1269 | BML1269 | 0.1534 | 0.8466 | 0.0246 | 0.103 | 0.8723 | 0.0002 | 0.4451 | 0.0476 | 0.5071 | 0 | 0.4403 | 0.035 | 0.4299 | 0.0948 | 0 | |||
| Y1216 | Y1216 | 0.2456 | 0.7544 | 0.1754 | 0.3081 | 0.5165 | 0.0794 | 0.3199 | 0.3155 | 0.2853 | 0.0008 | 0.2404 | 0.3154 | 0.072 | 0.2152 | 0.1562 | |||
| LM-6 | LM-6 | 0.4432 | 0.5568 | 0.3845 | 0.2577 | 0.3577 | 0.3391 | 0.1772 | 0.2587 | 0.225 | 0.1184 | 0.1052 | 0.2484 | 0.03 | 0.0862 | 0.4118 | |||
| 10GY6057 | 10GY6057-2 | 0.1042 | 0.8958 | 0.0616 | 0.8118 | 0.1266 | 0.002 | 0.1631 | 0.8111 | 0.0238 | 0.0036 | 0.1588 | 0.8054 | 0.0242 | 0.0072 | 0.0008 | |||
| BML1243 | BML1243 | 0.3012 | 0.6988 | 0.219 | 0.202 | 0.5789 | 0.1188 | 0.3422 | 0.2126 | 0.3264 | 0.0006 | 0.2458 | 0.2104 | 0.0502 | 0.2831 | 0.21 | |||
| BML1234 | BML1234 | 0.82 | 0.18 | 0.7848 | 0.0768 | 0.1384 | 0.6719 | 0.2533 | 0.0744 | 0.0004 | 0.651 | 0.2313 | 0.0666 | 0.0004 | 0.0004 | 0.0502 | |||
| Y1224 | Y1224 | 0.754 | 0.246 | 0.705 | 0.0578 | 0.2372 | 0.649 | 0.1646 | 0.0572 | 0.1292 | 0.5679 | 0.114 | 0.0558 | 0.0032 | 0.1654 | 0.0938 | |||
| 1217 8107 | 1217 8107 | 0.2984 | 0.7016 | 0.2328 | 0.4902 | 0.277 | 0.1618 | 0.233 | 0.4849 | 0.1204 | 0.1522 | 0.2294 | 0.4622 | 0.1518 | 0 | 0.0044 | |||
| BML1228 | BML1228 | 0.572 | 0.428 | 0.5187 | 0.2866 | 0.1947 | 0.4517 | 0.1884 | 0.2895 | 0.0704 | 0.2831 | 0.1182 | 0.2727 | 0.0008 | 0.004 | 0.3213 | |||
| 08WSC166 | 08WSC166-1211 | 0.3796 | 0.6204 | 0.3104 | 0.2478 | 0.4418 | 0.2162 | 0.3099 | 0.2581 | 0.2158 | 0.0448 | 0.2074 | 0.2529 | 0.0024 | 0.2088 | 0.2837 | |||
| LX350 | LX350 | 0.279 | 0.721 | 0.2194 | 0.374 | 0.4066 | 0.247 | 0.0228 | 0.3624 | 0.3678 | 0.059 | 0.0006 | 0.3473 | 0.2009 | 0.041 | 0.3513 | |||
| Y1217 | Y1217 | 0.386 | 0.614 | 0.3308 | 0.3378 | 0.3314 | 0.2773 | 0.1992 | 0.3397 | 0.1838 | 0.238 | 0.1574 | 0.3349 | 0.0956 | 0.092 | 0.082 | |||
| XBY2193 | SUWAN germplasm | 0.9438 | 0.0562 | 0.924 | 0.028 | 0.048 | 0.93 | 0.0022 | 0.0128 | 0.055 | 0.9021 | 0.0006 | 0.0172 | 0.0452 | 0.0092 | 0.0256 | |||
| 10GY92-121 | 10GY92-121 | 0.9506 | 0.0494 | 0.9158 | 0.0016 | 0.0826 | 0.888 | 0.0624 | 0.0006 | 0.049 | 0.8441 | 0.0174 | 0.0002 | 0.0004 | 0.1371 | 0.0008 | |||
| 10GY76-111 | 10GY76-111 | 0.9478 | 0.0522 | 0.9284 | 0.0508 | 0.0208 | 0.9286 | 0.003 | 0.0422 | 0.0262 | 0.915 | 0.0002 | 0.0292 | 0.0074 | 0.048 | 0.0002 | |||
| U8112 | Hybrid3382 | 0.0256 | 0.9744 | 0 | 0.0008 | 0.9992 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0.4464 | 0.5536 | 0 | |||
| W031-1 | W031-1 | 0.566 | 0.434 | 0.4721 | 0.0014 | 0.5265 | 0.461 | 0.0982 | 0 | 0.4408 | 0.3887 | 0.0498 | 0.0002 | 0.2238 | 0.2535 | 0.084 | |||
| C2010-3 | C2010-3 | 0.169 | 0.831 | 0.122 | 0.6961 | 0.182 | 0.1088 | 0.0596 | 0.6939 | 0.1376 | 0.0348 | 0.0108 | 0.6935 | 0.0104 | 0.113 | 0.1374 | |||
| FG-1 | Tropical germplasm | 0.991 | 0.009 | 0.9678 | 0.0024 | 0.0298 | 0.9322 | 0.066 | 0.0006 | 0.0012 | 0.9286 | 0.0536 | 0.001 | 0.0004 | 0.015 | 0.0014 | |||
| LS-22 | LS-22 | 0.6272 | 0.3728 | 0.5636 | 0.0404 | 0.396 | 0.5355 | 0.1407 | 0.0346 | 0.2892 | 0.2858 | 0.0376 | 0.028 | 0.0004 | 0.2604 | 0.3878 | |||
| CD30M | Hybrid CGT-15 | 0.555 | 0.445 | 0.5101 | 0.4021 | 0.0878 | 0.3649 | 0.2408 | 0.3943 | 0 | 0.3666 | 0.212 | 0.3892 | 0 | 0.0314 | 0.0008 | |||
| LZM009 | Tropical germplasm | 0.39 | 0.61 | 0.3398 | 0.549 | 0.1112 | 0.3182 | 0.0722 | 0.5452 | 0.0644 | 0.203 | 0.0044 | 0.5355 | 0.0004 | 0.0554 | 0.2012 | |||
| XH05 | Yunnan White XH05 × American hybrid | 0.4842 | 0.5158 | 0.4317 | 0.3377 | 0.2306 | 0.4253 | 0.0602 | 0.335 | 0.1795 | 0.2417 | 0 | 0.3269 | 0.0004 | 0.2717 | 0.1593 | |||
| 06WAM210 | CIMMYT line | 0.868 | 0.132 | 0.8272 | 0.0016 | 0.1712 | 0.814 | 0.0504 | 0.0006 | 0.135 | 0.6798 | 0.0006 | 0.0002 | 0.001 | 0.1043 | 0.2141 | |||
| 06WAM110 | CIMMYT line | 0.9992 | 0.0008 | 0.9976 | 0.0024 | 0 | 0.999 | 0.0002 | 0.0004 | 0.0004 | 0.9906 | 0 | 0.0008 | 0.0004 | 0.0012 | 0.007 | |||
| GCML32 | CIMMYT line | 0.8922 | 0.1078 | 0.8624 | 0.0374 | 0.1002 | 0.8679 | 0.0066 | 0.0272 | 0.0982 | 0.7562 | 0 | 0.0214 | 0.0062 | 0.0556 | 0.1606 | |||
| GCML57 | CIMMYT line | 0.934 | 0.066 | 0.902 | 0.0186 | 0.0794 | 0.8398 | 0.1388 | 0.0162 | 0.0052 | 0.8172 | 0.1258 | 0.0138 | 0.0018 | 0.0062 | 0.0352 | |||
| GCML140 | CIMMYT line | 0.8944 | 0.1056 | 0.8613 | 0.053 | 0.0858 | 0.7979 | 0.1447 | 0.0524 | 0.005 | 0.793 | 0.137 | 0.0504 | 0.0138 | 0.0028 | 0.003 | |||
| GCML152 | CIMMYT line | 0.954 | 0.046 | 0.932 | 0.0062 | 0.0618 | 0.932 | 0.0012 | 0.0012 | 0.0656 | 0.8449 | 0 | 0.0004 | 0.0074 | 0.0012 | 0.1461 | |||
| GCML157 | CIMMYT line | 0.988 | 0.012 | 0.974 | 0.0234 | 0.0026 | 0.9762 | 0.002 | 0.0124 | 0.0094 | 0.9424 | 0.0006 | 0.0042 | 0.0124 | 0.0002 | 0.0402 | |||
| MX714 | YA3237 / CIMMYT hybrid | 0.74 | 0.26 | 0.688 | 0.0024 | 0.3096 | 0.6703 | 0.09 | 0.0006 | 0.239 | 0.5599 | 0.0524 | 0.0004 | 0.1184 | 0.0282 | 0.2408 | |||
| DH40 | DH40 | 0.6294 | 0.3706 | 0.5657 | 0.0032 | 0.4311 | 0.567 | 0.0748 | 0.0006 | 0.3575 | 0.1595 | 0 | 0.0006 | 0.0044 | 0.1243 | 0.7112 | |||
| ZP2012 | Derived from Zhengda hybrid | 0.3112 | 0.6888 | 0.2525 | 0.5663 | 0.1812 | 0.0748 | 0.3517 | 0.5731 | 0.0004 | 0.0774 | 0.3513 | 0.5642 | 0 | 0.0068 | 0.0002 | |||
| P44 | P44 | 0.455 | 0.545 | 0.4069 | 0.4033 | 0.1898 | 0.3972 | 0.0652 | 0.401 | 0.1365 | 0.2531 | 0 | 0.3921 | 0 | 0.2903 | 0.0646 | |||
| 65232B | 6237 × Shen5003 | 0.348 | 0.652 | 0.2627 | 0.0492 | 0.6881 | 0.3165 | 0.0028 | 0.0134 | 0.6673 | 0.1757 | 0 | 0.0002 | 0.5612 | 0 | 0.2628 | |||
| CLRCW48 | CIMMYT line | 0.8928 | 0.1072 | 0.8614 | 0.0142 | 0.1244 | 0.8615 | 0.0246 | 0.006 | 0.108 | 0.7573 | 0.0012 | 0.005 | 0.0162 | 0.051 | 0.1693 | |||
| CLWN201 | CIMMYT line | 0.912 | 0.088 | 0.8794 | 0.0132 | 0.1074 | 0.8624 | 0.0578 | 0.0044 | 0.0754 | 0.7984 | 0.0398 | 0.0032 | 0.026 | 0.0376 | 0.095 | |||
| CLWN205 | CIMMYT line | 0.9692 | 0.0308 | 0.9266 | 0.0016 | 0.0718 | 0.9266 | 0.0058 | 0.0002 | 0.0674 | 0.833 | 0.0006 | 0.0002 | 0.0234 | 0.0018 | 0.141 | |||
| CLWN226 | CIMMYT line | 0.9062 | 0.0938 | 0.8734 | 0.0188 | 0.1078 | 0.8674 | 0.0386 | 0.0084 | 0.0856 | 0.788 | 0.015 | 0.0082 | 0.0234 | 0.0412 | 0.1242 | |||
| CLWN227 | CIMMYT line | 0.9374 | 0.0626 | 0.9044 | 0.0058 | 0.0898 | 0.8934 | 0.031 | 0.0008 | 0.0748 | 0.7835 | 0.0036 | 0.0014 | 0.0138 | 0.003 | 0.1947 | |||
| CLWN247 | CIMMYT line | 0.908 | 0.092 | 0.8758 | 0.0146 | 0.1096 | 0.8826 | 0.0112 | 0.0044 | 0.1018 | 0.7786 | 0.0006 | 0.0042 | 0.012 | 0.0626 | 0.142 | |||
| CLWN250 | CIMMYT line | 0.9598 | 0.0402 | 0.9227 | 0.0016 | 0.0757 | 0.9224 | 0.0052 | 0.0006 | 0.0718 | 0.8406 | 0.0006 | 0.0002 | 0.033 | 0.0038 | 0.1218 | |||
| CLWN251 | CIMMYT line | 0.9816 | 0.0184 | 0.941 | 0.0016 | 0.0574 | 0.939 | 0.003 | 0.0002 | 0.0578 | 0.8518 | 0.0006 | 0.0002 | 0.006 | 0.011 | 0.1304 | |||
| CL02450 | CIMMYT line | 0.935 | 0.065 | 0.9128 | 0.0212 | 0.066 | 0.9106 | 0.0186 | 0.009 | 0.0618 | 0.808 | 0.0006 | 0.007 | 0.0016 | 0.0358 | 0.147 | |||
| CL02603 | CIMMYT line | 0.8988 | 0.1012 | 0.871 | 0.0496 | 0.0794 | 0.8698 | 0.0168 | 0.0414 | 0.072 | 0.7909 | 0.0012 | 0.036 | 0.023 | 0.0318 | 0.1171 | |||
| CL02720 | CIMMYT line | 0.9534 | 0.0466 | 0.9244 | 0.006 | 0.0696 | 0.9192 | 0.0216 | 0.0014 | 0.0578 | 0.8128 | 0.0006 | 0.0012 | 0.0046 | 0.002 | 0.1788 | |||
| CLRCY034 | CIMMYT line | 0.9234 | 0.0766 | 0.893 | 0.0098 | 0.0972 | 0.8731 | 0.0576 | 0.0026 | 0.0668 | 0.7935 | 0.0428 | 0.002 | 0.014 | 0.0096 | 0.1381 | |||
| CLRCY041 | CIMMYT line | 0.955 | 0.045 | 0.9338 | 0.0152 | 0.051 | 0.9254 | 0.0336 | 0.006 | 0.035 | 0.8437 | 0.0038 | 0.003 | 0.0004 | 0.0316 | 0.1175 | |||
| CLYN214 | CIMMYT line | 0.9946 | 0.0054 | 0.9698 | 0.0016 | 0.0286 | 0.968 | 0.001 | 0 | 0.031 | 0.8846 | 0 | 0.0002 | 0.0014 | 0.0004 | 0.1134 | |||
| CLYN223 | CIMMYT line | 0.9532 | 0.0468 | 0.918 | 0.0024 | 0.0796 | 0.8986 | 0.0462 | 0.0006 | 0.0546 | 0.8109 | 0.0262 | 0.0004 | 0.0012 | 0.0258 | 0.1355 | |||
| CML268 | CIMMYT line | 0.9072 | 0.0928 | 0.8794 | 0.0306 | 0.09 | 0.8837 | 0.0092 | 0.0164 | 0.0907 | 0.8136 | 0.0012 | 0.0086 | 0.0432 | 0.0098 | 0.1235 | |||
| CML282 | CIMMYT line | 0.9502 | 0.0498 | 0.921 | 0.0024 | 0.0766 | 0.915 | 0.0138 | 0.0006 | 0.0706 | 0.8228 | 0.0006 | 0.0006 | 0.0018 | 0.0486 | 0.1256 | |||
| CML291 | CIMMYT line | 0.91 | 0.09 | 0.8842 | 0.0284 | 0.0874 | 0.884 | 0.0098 | 0.02 | 0.0862 | 0.8138 | 0.0006 | 0.0178 | 0.0202 | 0.0508 | 0.0968 | |||
| CML308 | CIMMYT line | 0.911 | 0.089 | 0.8864 | 0.0356 | 0.078 | 0.8842 | 0.0224 | 0.0284 | 0.065 | 0.8088 | 0.0016 | 0.0258 | 0.0048 | 0.049 | 0.11 | |||
| CML379 | CIMMYT line | 0.9392 | 0.0608 | 0.9022 | 0.0024 | 0.0954 | 0.905 | 0.0072 | 0.0006 | 0.0872 | 0.778 | 0 | 0.0002 | 0.0024 | 0.0426 | 0.1767 | |||
| CML447 | CIMMYT line | 0.9124 | 0.0876 | 0.881 | 0.0254 | 0.0936 | 0.8832 | 0.0086 | 0.012 | 0.0962 | 0.7772 | 0.0006 | 0.0096 | 0.0208 | 0.0158 | 0.176 | |||
| CML451 | CIMMYT line | 0.964 | 0.036 | 0.9328 | 0.0024 | 0.0648 | 0.9312 | 0.0054 | 0.0006 | 0.0628 | 0.86 | 0.0008 | 0.0004 | 0.0126 | 0.0324 | 0.0938 | |||
| TL98A1709-20 | CIMMYT line | 0.7532 | 0.2468 | 0.7071 | 0.036 | 0.2569 | 0.6789 | 0.116 | 0.0304 | 0.1748 | 0.4937 | 0.044 | 0.027 | 0.0022 | 0.1096 | 0.3235 | |||
| TL96B | CIMMYT line | 0.716 | 0.284 | 0.6706 | 0.0544 | 0.275 | 0.6453 | 0.107 | 0.0494 | 0.1984 | 0.5074 | 0.0524 | 0.0482 | 0.0352 | 0.1427 | 0.2141 | |||
| 98WV9 | CIMMYT line | 0.943 | 0.057 | 0.9126 | 0.0026 | 0.0848 | 0.904 | 0.0262 | 0.0006 | 0.0692 | 0.8074 | 0.0006 | 0.0004 | 0.001 | 0.0531 | 0.1375 | |||
| SW01D1031-14 | CIMMYT line | 0.931 | 0.069 | 0.9094 | 0.0414 | 0.0492 | 0.912 | 0.0034 | 0.0286 | 0.056 | 0.8724 | 0.0006 | 0.0268 | 0.035 | 0.0048 | 0.0604 | |||
| SW01D1031-17 | CIMMYT line | 0.915 | 0.085 | 0.8908 | 0.0266 | 0.0826 | 0.8857 | 0.0236 | 0.016 | 0.0748 | 0.8426 | 0.0096 | 0.0136 | 0.048 | 0.0028 | 0.0834 | |||
| SW01D1058-5 | CIMMYT line | 0.956 | 0.044 | 0.9396 | 0.0366 | 0.0238 | 0.9422 | 0.0032 | 0.0246 | 0.03 | 0.9024 | 0.0006 | 0.021 | 0.0126 | 0.0018 | 0.0616 | |||
| SW01D1058-2 | CIMMYT line | 0.956 | 0.044 | 0.9392 | 0.0362 | 0.0246 | 0.9428 | 0.0028 | 0.0238 | 0.0306 | 0.9028 | 0.0006 | 0.0204 | 0.0128 | 0.0022 | 0.0612 | |||
| SW01D1058-7 | CIMMYT line | 0.8398 | 0.1602 | 0.8098 | 0.0616 | 0.1286 | 0.7981 | 0.041 | 0.0504 | 0.1106 | 0.6955 | 0.0168 | 0.042 | 0.0538 | 0.0002 | 0.1918 | |||
| BANTAN2003 | CIMMYT line | 0.9238 | 0.0762 | 0.8579 | 0.0008 | 0.1413 | 0.8418 | 0.0538 | 0 | 0.1044 | 0.7271 | 0.0166 | 0.0002 | 0.0102 | 0.0628 | 0.1832 | |||
| CG698C102 | 698-3 improved line | 0.3452 | 0.6548 | 0.2332 | 0.0654 | 0.7014 | 0.0492 | 0.6192 | 0.0788 | 0.2527 | 0 | 0.4894 | 0.0016 | 0.0004 | 0.0986 | 0.41 | |||
| CG921 | Hai921 improved line | 0.316 | 0.684 | 0.2659 | 0.4825 | 0.2517 | 0.2384 | 0.1122 | 0.4785 | 0.1708 | 0.0504 | 0.0408 | 0.4703 | 0.0046 | 0.0802 | 0.3537 | |||
| CG178 | 178 improved line | 0.3504 | 0.6496 | 0.3012 | 0.5536 | 0.1452 | 0.3092 | 0.001 | 0.5415 | 0.1483 | 0.1456 | 0 | 0.5354 | 0.0014 | 0.0926 | 0.225 | |||
| Lian87 | Lian87 | 0.624 | 0.376 | 0.5663 | 0.004 | 0.4297 | 0.5534 | 0.1026 | 0.0006 | 0.3434 | 0.2204 | 0.0078 | 0.0002 | 0.099 | 0.023 | 0.6496 | |||
| CP16 | Unknown | 0.4032 | 0.5968 | 0.3378 | 0.2189 | 0.4433 | 0.322 | 0.1232 | 0.216 | 0.3388 | 0 | 0.0006 | 0.1632 | 0.0376 | 0.0012 | 0.7974 | |||
| K305 | American hybrid 2021 | 0.207 | 0.793 | 0.1214 | 0.1036 | 0.775 | 0.1692 | 0.0012 | 0.0794 | 0.7502 | 0 | 0 | 0.0002 | 0.4727 | 0 | 0.5271 | |||
| 811 | Derived from 136-87mutant | 0.099 | 0.901 | 0.0468 | 0.6062 | 0.347 | 0.0784 | 0.004 | 0.5953 | 0.3223 | 0 | 0 | 0.5514 | 0.1252 | 0.0002 | 0.3232 | |||
| K363 | 48-2 × 156 | 0.4088 | 0.5912 | 0.3246 | 0.1536 | 0.5218 | 0.2806 | 0.1972 | 0.1526 | 0.3696 | 0.0988 | 0.0888 | 0.1584 | 0.0642 | 0.3371 | 0.2527 | |||
| K102 | K102 | 0.3828 | 0.6172 | 0.3214 | 0.409 | 0.2696 | 0.195 | 0.3129 | 0.4191 | 0.073 | 0.0508 | 0.2489 | 0.4006 | 0.0004 | 0.0532 | 0.2461 | |||
| S0035 | 78599 | 0.5092 | 0.4908 | 0.4238 | 0.0404 | 0.5358 | 0.3652 | 0.2172 | 0.0386 | 0.379 | 0.2212 | 0.1572 | 0.0316 | 0.179 | 0.145 | 0.266 | |||
| B0069 | Shuangyu2 male parent | 0.2348 | 0.7652 | 0.1763 | 0.5004 | 0.3233 | 0.1596 | 0.0978 | 0.4961 | 0.2464 | 0.0961 | 0.0518 | 0.4889 | 0.1425 | 0.1193 | 0.1013 | |||
| 81565 | Miandan 8 parent Mobai | 0.5712 | 0.4288 | 0.4977 | 0.004 | 0.4983 | 0.4824 | 0.1335 | 0.0012 | 0.3828 | 0.1344 | 0.005 | 0.0004 | 0.0152 | 0.2084 | 0.6365 | |||
| LYC-1 | LONG Xichang | 0.8442 | 0.1558 | 0.8166 | 0.1164 | 0.067 | 0.7856 | 0.085 | 0.1138 | 0.0156 | 0.7102 | 0.0544 | 0.1012 | 0.0012 | 0.019 | 0.114 | |||
| WZ-1 | W1 Wanzhou | 0.395 | 0.605 | 0.3503 | 0.4265 | 0.2232 | 0.3239 | 0.1044 | 0.4279 | 0.1438 | 0.2065 | 0.0384 | 0.4259 | 0.0018 | 0.1255 | 0.2019 | |||
| PB80 | 1067-1 × B73 / B73Ht.1 BC6, SS | 0.3164 | 0.6836 | 0.1303 | 0.0008 | 0.8689 | 0.1369 | 0.1361 | 0 | 0.727 | 0 | 0 | 0 | 0 | 1 | 0 | |||
| 787 | VA17 / VA29, NSS | 0.5922 | 0.4078 | 0.5257 | 0.0492 | 0.4251 | 0.5024 | 0.1326 | 0.0442 | 0.3208 | 0.228 | 0.0336 | 0.0406 | 0.014 | 0.174 | 0.5098 | |||
| 793 | 235 / B73, SS | 0.3612 | 0.6388 | 0.1758 | 0.0002 | 0.824 | 0.1645 | 0.1859 | 0 | 0.6497 | 0.0244 | 0.0108 | 0 | 0 | 0.961 | 0.0038 | |||
| W8034 | B14A/2 × B73, SS | 0.3372 | 0.6628 | 0.1718 | 0.0008 | 0.8274 | 0.1712 | 0.1278 | 0 | 0.7009 | 0 | 0 | 0 | 0 | 1 | 0 | |||
| PHV63 | 555 × Zap<4CB, NSS | 0.427 | 0.573 | 0.3454 | 0.1128 | 0.5418 | 0.3323 | 0.1264 | 0.1042 | 0.4371 | 0.136 | 0.0336 | 0.1012 | 0.1188 | 0.3323 | 0.2781 | |||
| PHW65 | (861 / 595) × 8211131X,NSS | 0.324 | 0.676 | 0.2525 | 0.3241 | 0.4235 | 0.2362 | 0.1236 | 0.3191 | 0.3211 | 0.0006 | 0.0122 | 0.2974 | 0.1177 | 0.0414 | 0.5308 | |||
| 2369 | 2702H × B73) × B73, SS | 0.29 | 0.71 | 0.112 | 0.0008 | 0.8872 | 0.1098 | 0.1536 | 0 | 0.7365 | 0 | 0 | 0.0002 | 0.1353 | 0.8645 | 0 | |||
| 87916W | B73 × W37-2 -B1-1-B1, SS | 0.3912 | 0.6088 | 0.267 | 0.0016 | 0.7314 | 0.2488 | 0.1562 | 0.0004 | 0.5946 | 0.1054 | 0.0212 | 0.0042 | 0.0506 | 0.7808 | 0.0378 | |||
| PHW52 | (B73 / G39) × 411242X, SS | 0.3292 | 0.6708 | 0.2152 | 0.0088 | 0.776 | 0.191 | 0.2136 | 0.0046 | 0.5908 | 0 | 0 | 0 | 0 | 1 | 0 | |||
| CA1108 | Huangzaosi improved line | 0.5522 | 0.4478 | 0.4892 | 0.1393 | 0.3715 | 0.4554 | 0.1572 | 0.1422 | 0.2452 | 0.2213 | 0.062 | 0.1395 | 0.0064 | 0.1589 | 0.4118 | |||
| Nan21-3 | Yogoslavic hybrid BC8241Ht | 0.6162 | 0.3838 | 0.5488 | 0.0074 | 0.4438 | 0.5069 | 0.1792 | 0.003 | 0.3109 | 0.1655 | 0.0782 | 0.0012 | 0.0022 | 0.1383 | 0.6146 | |||
| PH6WC | PH01N × PH09B, Reid population | 0.3422 | 0.6578 | 0.2371 | 0.0656 | 0.6973 | 0.2046 | 0.2074 | 0.0634 | 0.5247 | 0 | 0 | 0 | 0 | 1 | 0 | |||
| PH4CV | PH7V0 × PHBE2,Lancaster population | 0.489 | 0.511 | 0.4161 | 0.1186 | 0.4653 | 0.3353 | 0.2555 | 0.115 | 0.2943 | 0.014 | 0.1641 | 0.0976 | 0.0514 | 0.0608 | 0.6122 | |||
| Xun9058 | 6JK × 8085 Thailand | 0 | 1 | 0 | 0.3165 | 0.6835 | 0 | 0 | 0.3071 | 0.6929 | 0 | 0 | 0.2846 | 0.7154 | 0 | 0 | |||
| LX9801 | 502 × H21 | 0.7854 | 0.2146 | 0.7028 | 0.0008 | 0.2964 | 0.6807 | 0.105 | 0 | 0.2142 | 0.3228 | 0.0012 | 0 | 0.0004 | 0.0312 | 0.6444 | |||
| C24 | C24 | 0.2144 | 0.7856 | 0.1526 | 0.6832 | 0.1642 | 0.0826 | 0.1858 | 0.6846 | 0.047 | 0.0902 | 0.1865 | 0.6787 | 0.0408 | 0.0036 | 0.0002 | |||
| C09-1 | 09-1 | 0.4892 | 0.5108 | 0.4197 | 0.1944 | 0.3859 | 0.3746 | 0.1798 | 0.1934 | 0.2522 | 0.1774 | 0.1042 | 0.1932 | 0.0206 | 0.1678 | 0.3369 | |||
| L2010-3 | L2010-3 | 0.903 | 0.097 | 0.8848 | 0.092 | 0.0232 | 0.8503 | 0.0636 | 0.0845 | 0.0016 | 0.8376 | 0.065 | 0.0812 | 0.0086 | 0.0018 | 0.0058 | |||
| ZNC-4 | Unknown | 0.4628 | 0.5372 | 0.3369 | 0.0008 | 0.6623 | 0.3017 | 0.1996 | 0 | 0.4987 | 0.0008 | 0.08 | 0.0002 | 0.096 | 0.2389 | 0.584 | |||
| Qi533 | Derived from P136 abnormal plant | 0.3766 | 0.6234 | 0.2749 | 0.0576 | 0.6675 | 0.0382 | 0.6623 | 0.0742 | 0.2254 | 0.0028 | 0.6309 | 0.036 | 0.2492 | 0 | 0.081 | |||
| L6201 | 08-641 improved line | 0.746 | 0.254 | 0.6957 | 0.0542 | 0.2501 | 0.6365 | 0.1729 | 0.0538 | 0.1367 | 0.554 | 0.1218 | 0.0538 | 0.0044 | 0.1678 | 0.0982 | |||
| LSC127 | LSC127 | 0.4322 | 0.5678 | 0.3566 | 0.0858 | 0.5576 | 0.3615 | 0.086 | 0.0772 | 0.4754 | 0.1777 | 0.0046 | 0.0686 | 0.2417 | 0.1577 | 0.3496 | |||
| W30 | K22 improved line | 0.2692 | 0.7308 | 0.1612 | 0.0264 | 0.8124 | 0.0294 | 0.4131 | 0.0162 | 0.5413 | 0.0388 | 0.3786 | 0.0004 | 0.5231 | 0.0002 | 0.0588 | |||
| B73 | BSSS C5 | 0.287 | 0.713 | 0.058 | 0 | 0.942 | 0.0564 | 0.1625 | 0 | 0.7811 | 0 | 0 | 0 | 0 | 1 | 0 | |||
| B047 | B047 | 0.0024 | 0.9976 | 0.0002 | 0.9982 | 0.0016 | 0.0004 | 0.0012 | 0.9964 | 0.002 | 0 | 0 | 0.985 | 0 | 0.015 | 0 | |||
| W3189 | 18-599 improved line | 0.138 | 0.862 | 0.0746 | 0.6418 | 0.2836 | 0.0604 | 0.0737 | 0.6393 | 0.2266 | 0.0006 | 0.0402 | 0.6273 | 0.1052 | 0.1054 | 0.1212 | |||
| 9782 | (48-2 × 5003)space induced mutant × foreign hybrid line | 0.12 | 0.88 | 0.0458 | 0.3899 | 0.5643 | 0.0812 | 0.007 | 0.3777 | 0.5341 | 0 | 0 | 0.3165 | 0.3247 | 0 | 0.3589 | |||
| 646 | Lancaster L3874 × Tropical germplasm | 0.9302 | 0.0698 | 0.908 | 0.0302 | 0.0618 | 0.9152 | 0.0028 | 0.0168 | 0.0652 | 0.8848 | 0.001 | 0.0218 | 0.0424 | 0.0258 | 0.0242 | |||
| CZ2142 | Unknown | 0.5928 | 0.4072 | 0.5228 | 0.0722 | 0.405 | 0.2716 | 0.5986 | 0.1026 | 0.0272 | 0.1966 | 0.5228 | 0.078 | 0.0004 | 0.0702 | 0.132 | |||
| CZ205-2 | Unknown | 0.4716 | 0.5284 | 0.3939 | 0.0968 | 0.5093 | 0.3563 | 0.1848 | 0.0932 | 0.3657 | 0.0154 | 0.0788 | 0.086 | 0.012 | 0.1787 | 0.6291 |
Genotype
A total of 56,110 SNPs on the MaizeSNP50 BeadChip were used for genotyping the panel. Detailed information for this chip can be downloaded from the Illumina MaizeSNP50 website and the position information of SNPs according to B73 RefGen_v2 can be downloaded from NCBI (National Center for Biotechnology Information) GEO website. One month after seed germination, eight leaf pieces from differentindividualsofthesameinbredlinewere placed into one tube, and two replicates were collected for DNA extraction and genotyping. Using Illumina GenomeStudio, a powerful genotyping tool, each SNP was rechecked manually to identify any errors following Yan. Then the raw file exported by GenomeStudio was transformed to PowerMarker V3.25 format for calculation of summary statistics. SNPs with missing rate >20 %, heterozygosity >20 % and minor allele frequency (MAF) '<'0.05 were excluded from the genotyping data, leaving 44,104 high-quality SNPs for further analysis.
After filtering of the total SNP data, 43,735 high-quality SNPs with identified physical position were used for summary description. In this subset of SNPs, the average marker density is approximately 52 kb per SNP, the average distance between adjacent SNPs is 47 kb, and the position of each SNP is listed in Additional file 2: Table S2. The MAF, GD, PIC value, and heterozygosity among chromosomes showed little variation among chromosomes by PowerMarker (Fig. 1). However, the numbers of SNPs showing significant variation, from 3128 on Chromosome (Chr) 10 to 6834 on Chr 1 were correlated with the lengths of the corresponding chromosomes. MAF was uniformly distributed from 0.1 to 0.5 among SNPs, but the trends of GD and PIC were similar, with most SNPs showing high levels of these parameters (48 % showing GD >0.45 and 55 % showing PIC >0.35).
Population structure and kinship
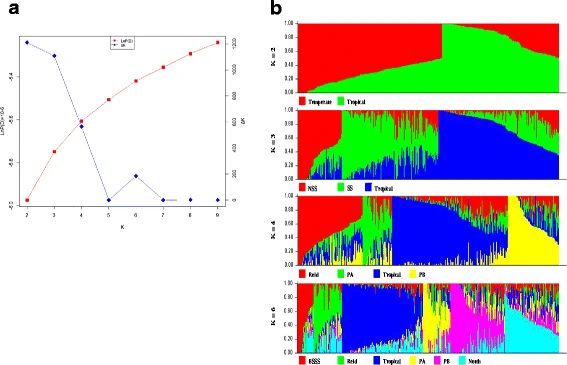
STRUCTURE software was used to identify population structure. Because LnP(D) continuously increased with the K value, it was not capable of identifying groups. Accordingly, the ΔK value was calculated for each K. The peaks of line plot suggested that the entire panel could be divided into two, three, four, six groups in order of possibility based on breeding experience and pedigree information. When K = 2, the whole panel could be grouped into Tropical and Temperate groups which the representative inbred lines were S37 and 18–599; When K = 3, the Temperate group was further divided into SS and NSS groups, including representative inbred lines Qi319 and Ye478, respectively; When K = 4, PA and PB were further separated which the representative inbred lines were 698–3 and 18–599; When K = 6, besides the Tropical subgroup, other inbred lines could be clustered into BSSS, Reid, PA, PB and North group (other temperate germplasms mostly derived from North China including Lancaster, SPT and LRC), the representative inbred lines were B73, Ye478, 698–3, 18599, and Dan340, respectively. These results were largely consistent with known pedigree information. Other lines with lower possibility were clustered into a mixed group. PCA analysis based on the results of STRUCTURE showed a clear structure separation of the panel. When K = 2, the PCA plot showed a clear separation of Temperate and Tropical groups (Fig. 3a). The mixed group was located between the other two groups. The Temperate group showed more dispersion than the Tropical group did, suggesting that the temperate lines could be divided into further subgroups. For K = 3, the NSS and SS groups were clearly separated from the Temperate group and the genetic distance between SS and NSS was less than that between NSS and Tropical (Fig. 3b). For K = 4, the PA group was located at the center, mixed with other groups, while the other groups showed relatively stable distribution (Fig. 3c). For K = 6, the PCA plot showed a complex distribution of subgroups, except for the PB and Tropical groups. The BSSS group was largely from the Reid group, and the PA and North groups located at the center, showed a complex source. These results also agreed with the conclusions from STRUCTURE that the groups were gradually separated based on one main group, Temperate, with K increasing and the Tropical group remaining relatively stable during this process. To confirm the separation between the groups from the Temperate group, PCA analysis was also performed after removal of the Tropical and Mixed groups for K = 3, K = 4, and K = 6. Clear separations of the Temperate group were showed for K = 3 and K = 4, but more complex clustering for K = 6.
Related Publication
Zhang, X., Zhang, H., Li, L. et al. Characterizing the population structure and genetic diversity of maize breeding germplasm in Southwest China using genome-wide SNP markers. BMC Genomics 17, 697 (2016). https://doi.org/10.1186/s12864-016-3041-3