MGTRP: Maize Genetic Transformation Resources Platform
## This platform provides the resources as follows:
- the phenotypes and genotypes of embryogenic callus induction and regeneration ability-related traits and Agrobacterium-mediated transformation abilities from about 700 inbred lines collected from China, USA, and Mexico etc. ;
- the reference genomes, full-length transcriptomes, and dynamic transcriptome of A188, 18-599R, and C01.
All the resources are freely available and openly accessed for the researchers worldwide.
Maize (Zea mays L.) is a primary global crop supplying the food, feed, and industrial materials industries. Genetic transformation is presently widely used to improve yield and stress resistance and for gene function validation in maize, which largely depend on callus induction and regeneration from maize immature embryos.
Phenotype
Different inbred lines showed great differences in embryogenic callus induction (Fig. 1A), green dot state (Fig. 1B) and green seedling regeneration (Fig. 1C).
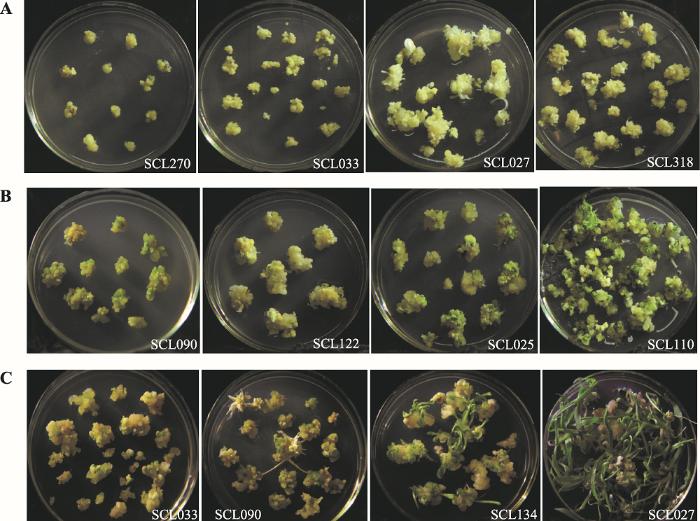
Table 1. Population characteristics of all traits in different environments
| Env | Traits | CCI1 | CCI2 | GCR | CDR | CPN | CRR | CBR |
|---|---|---|---|---|---|---|---|---|
| XSBN1 | Mean | - | - | 42.4642 | 19.5236 | 1.3766 | 8.2896 | 36.6837 |
| SD | - | - | 23.4446 | 17.9975 | 0.65 | 10.3111 | 26.955 | |
| Variance | - | - | 549.649 | 323.909 | 0.422 | 106.319 | 726.57 | |
| Skewness | - | - | -0.357 | 0.7 | 3.474 | 1.096 | 0.637 | |
| Kurtosis | - | - | -0.47 | 0.202 | 17.375 | 0.25 | -0.502 | |
| Min | - | - | 0 | 0 | 1 | 0 | 0 | |
| Max | - | - | 90 | 83.845 | 5.5955 | 38.4299 | 90 | |
| CZ | Mean | 1.9122 | 1.5065 | 38.0679 | 16.6353 | 1.2468 | 14.6485 | 42.1359 |
| SD | 0.4336 | 0.1911 | 26.6835 | 20.4784 | 0.493 | 17.9865 | 28.8642 | |
| Variance | 0.188 | 0.0365 | 712.0103 | 419.3666 | 0.2431 | 323.5139 | 833.1405 | |
| Skewness | 2.0167 | -0.0626 | 0.2101 | 1.63 | 2.8373 | 1.8715 | 0.373 | |
| Kurtosis | 6.784 | 0.5188 | -0.8583 | 2.6546 | 8.188 | 4.8099 | -1.0878 | |
| Min | 1.1256 | 1 | 0 | 0 | 1 | 0 | 0 | |
| Max | 3.9964 | 2.0351 | 90 | 90 | 3.4624 | 90 | 90 | |
| XSBN2 | Mean | 1.976 | 1.5378 | 48.959 | 15.7448 | 1.2106 | 11.563 | 34.2689 |
| SD | 0.5563 | 0.3233 | 24.7053 | 14.0858 | 0.3766 | 14.4439 | 25.1472 | |
| Variance | 0.31 | 0.105 | 610.351 | 198.409 | 0.142 | 208.626 | 632.383 | |
| Skewness | 0.466 | 0.578 | -0.505 | 0.677 | 2.435 | 1.866 | 0.865 | |
| Kurtosis | 2.092 | 1.887 | -0.589 | 0.029 | 5.551 | 4.743 | -0.195 | |
| Min | 1 | 1 | 0 | 0 | 1 | 0 | 0 | |
| Max | 4.1473 | 2.6614 | 90 | 60.9829 | 2.6877 | 75.9638 | 90 |
IBM Genotype

A total of 1,151,856 high-quality SNPs were generated from the parental SNP set. In total, 76.5 % of the SNPs (1,505,749 SNPs) were verified at least once from the 280 lines in the Syn10 population, and 23.5 % of the SNPs were excluded because no SNPs were successfully validated from the Syn10 population. In our database, 18.77 % (216,254 out of 1,151,856 SNPs) were verified 1–4 times in the Syn10 population. Additionally, 81.23 % (935,602 of the verified high-quality SNPs) were verified more than five times in the Syn10 population. Finally, a total of 2,961 SSR, RFLP, and IDP markers with precise physical coordinates as well as extra 6,618 bin markers and 1,151,856 SNPs were collected to create a high-quality and density-integrated map.
A high-density genetic map was constructed for the IBM Syn10 population with all 6,618 recombination bin markers. The map quality was reliable as only adjacent markers showed strong linkage with 11,198.5 cM genetic distance in the map (Additional file 9). The average genetic distance of the bin markers was 1.7 cM, which represented an increase in the marker density compared with the 4.7 cM observed between the adjacent markers, for a total of 6,242.7 cM with 1,340 markers in the Syn4 population.
Common features, such as the recombination rate and genetic vs. physical distance were compared between the Syn4 and Syn10 populations. Figure 3 shows the relationship between the genetic and physical positions on all chromosomes between the two populations. The recombination suppression was observed near the centromeres, and higher recombination rates were found to be predominant near both telomeres. The average ratio of genetic-to-physical distance was 6.95 cM/Mb for the whole genome, which is higher than that of the Syn4 population (4.55 cM/Mb).


The genotype of IBM can be download here: [ 12864_2015_2242_MOESM13_ESM.xlsx ] .
GWAS Genotype
A total of 56,110 SNPs on the MaizeSNP50 BeadChip were used for genotyping the panel. Detailed information for this chip can be downloaded from the Illumina MaizeSNP50 website and the position information of SNPs according to B73 RefGen_v2 can be downloaded from NCBI (National Center for Biotechnology Information) GEO website. One month after seed germination, eight leaf pieces from differentindividualsofthesameinbredlinewere placed into one tube, and two replicates were collected for DNA extraction and genotyping. Using Illumina GenomeStudio, a powerful genotyping tool, each SNP was rechecked manually to identify any errors following Yan. Then the raw file exported by GenomeStudio was transformed to PowerMarker V3.25 format for calculation of summary statistics. SNPs with missing rate >20 %, heterozygosity >20 % and minor allele frequency (MAF) '<'0.05 were excluded from the genotyping data, leaving 44,104 high-quality SNPs for further analysis.
The genotype of GWAS can be download here: Genotye.hmp.txt .
The SNP information of GWAS can be download here: SNP_56110_information.txt .
Please contact us for the file download code.
Callus Induction Rate (CIR)
The callus induction rate was defined as: callus induction rate (%) = (number of explants produced callus/total number of explants cultured) × 100%
ZmSAUR15 affects callus formation in maize
The ability of immature maize (Zea mays) embryos to form embryonic calluses (ECs) is highly genotype dependent, which limits transgenic breeding development in maize. Here, we report the association map-based cloning of ZmSAUR15 using an association panel (AP) consisting of 309 inbred lines with diverse formation abilities for ECs. We demonstrated that ZmSAUR15, which encodes a small auxin-upregulated RNA, acts as a negative effector in maize EC induction. Polymorphisms in the ZmSAUR15 promoter that influence the expression of ZmSAUR15 transcripts modulate the EC induction capacity in maize. ZmSAUR15 is involved in indole-3-acetic acid biosynthesis and cell division in immature embryo-derived callus. The ability of immature embryos to induce EC formation can be improved by the knockout of ZmSAUR15, which consequently increases the callus regeneration efficiency. Our study provides new insights into overcoming the genotypic limitations associated with EC formation and improving genetic transformation in maize.
Phenotype
The AP, consisting of 309 maize inbred lines, was grown in three locations. The immature embryos of the AP collected from each location were used to investigate the ECIR phenotype. The descriptive analysis results showed that the average ECIR of the AP was 5.60, 3.15 and 2.03% in the locations Chongzhou (CZ), Xishuangbanna (XSBN) and Chengdu (CD), respectively. The highest variation in the ECIR was observed in XSBN, ranging from 0 to 73.33% (Table 1). Analysis of variance (ANOVA) results showed that the ECIR was significantly affected by genotype, environment and the interaction between both. The H2 estimate of the ECIR was 53.97%, indicating that ECIR is mainly controlled by genetic factors in maize.
Table. Analysis of variance of the callus traits in the three environments
| Trait | Source of variation | Mean square | df | F | Significance | H2 |
|---|---|---|---|---|---|---|
| ECIR | Intercept | 23 390.265 | 1 | 34.859 | <0.0001 | 53.97% |
| Genotype (G) | 278.618 | 308 | 48.555 | <0.0001 | ||
| Environment (E) | 1614.077 | 2 | 281.288 | <0.0001 | ||
| G × E | 128.235 | 383 | 22.348 | <0.0001 | ||
| Residual error | 5.738 | 1216 | <0.0001 |
ECIR, embryonic callus induction rate; H2 , broad-sense heritability.
Variants in the CDS and promoter of ZmSAUR15
To determine which gene(s) in the auxin signal transduction pathway affect maize EC initiation, we used high-ECIR lines and low-ECIR lines to amplify the CDS and promoter sequences of the 16 auxin signal-related genes identified as responding to 2,4-D in our previous study. No significant genetic variation was observed for any of the genes between the two groups with ECIR disparity except for ZmSAUR15, the CDS and promoter of which displayed genetic discrepancies between the contrasting groups. To further confirm the genetic variants in ZmSAUR15 associated with the ECIR, sequences covering the entire CDS of ZmSAUR15 (429–444 bp) were amplified from the cDNAs of the 309 lines. A total of 25 SNPs and 14 InDels with minor allele frequency (MAF) ≥ 0.05 were identified. Association analysis using a general linear model (GLM) and a mixed linear model (MLM) indicated that 19 SNPs and five InDels were significantly (P ≤ 0.05) associated with the ECIR of maize in the three environments and using the best linear unbiased prediction (BLUP) values. All the loci were repeatedly detected across at least two environments and in both models.
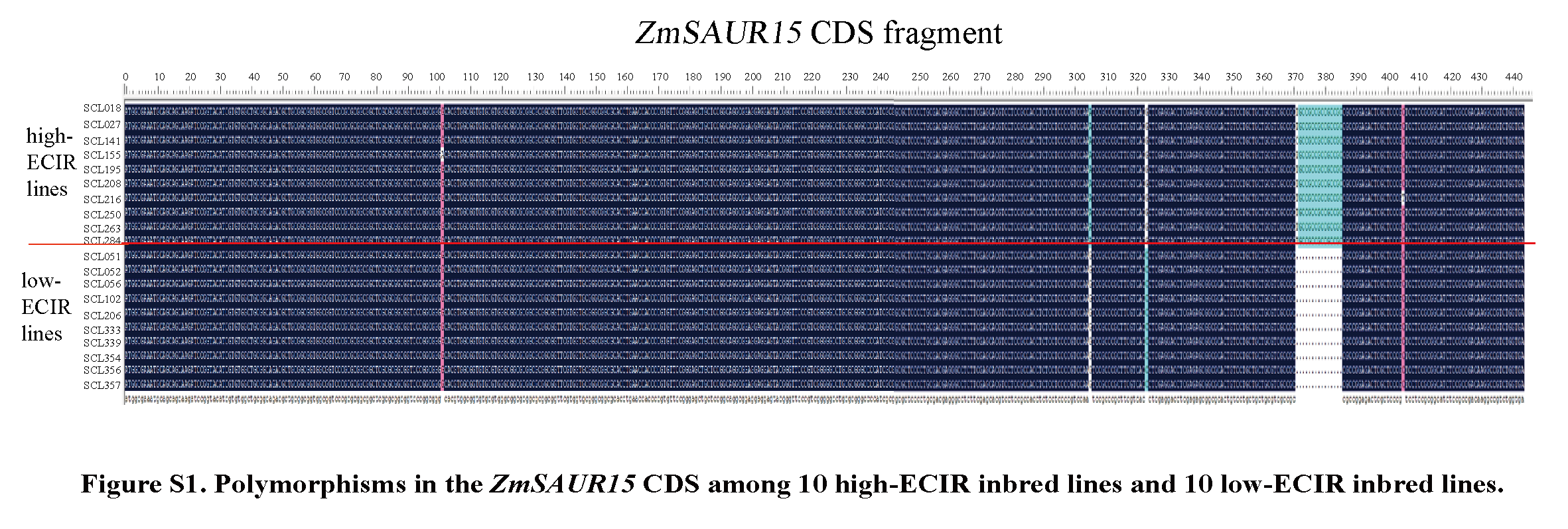
These 24 variants classified these lines into three main haplotypes, Hap1, Hap2 and Hap3, which contained 94, 60 and 129 lines, espectively. ANOVA results revealed that there was a significant (**P < 0.01) difference in the ECIR (average among the three environments) between the haplotypes; Hap2 and Hap1 (represented by SCL155 and B73, respectively) displayed the highest and lowest ECIRs, respectively. Therefore, in this study, Hap2 and Hap1 were considered as the superior and inferior haplotypes for the ZmSAUR15 CDS, respectively.

Related Publication
- Zhang Xiaoling,Long Yun,Ge Fei,Guan Zhongrong,Zhang Xiaoxiang,Wang Yanli,Shen Yaou,Pan Guangtang. A genetic study of the regeneration capacity of embryonic callus from the maize immature embryo culture[J]. Hereditas(Beijing), 2017, 39(2): 143-155.
- Liu, H., Niu, Y., Gonzalez-Portilla, P.J. et al. An ultra-high-density map as a community resource for discerning the genetic basis of quantitative traits in maize. BMC Genomics 16, 1078 (2015). https://doi.org/10.1186/s12864-015-2242-5
- Wang, Y., He, S., Long, Y., Zhang, X., Zhang, X., Hu, H., Li, Z., Hou, F., Ge, F., Gao, S., Pan, G., Ma, L. and Shen, Y. (2022), Genetic variations in ZmSAUR15 contribute to the formation of immature embryo-derived embryonic calluses in maize. Plant J, 109: 980-991. https://doi.org/10.1111/tpj.15609